Landscape of potentially targetable receptor tyrosine kinase fusions in diverse cancers by DNA-based profiling
- PMID: 36369474
- PMCID: PMC9652465
- DOI: 10.1038/s41698-022-00325-0
Landscape of potentially targetable receptor tyrosine kinase fusions in diverse cancers by DNA-based profiling
Abstract
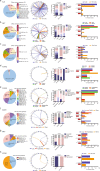
Recurrent fusions of receptor tyrosine kinases (RTKs) are often driving events in tumorigenesis that carry important diagnostic value and are potentially targetable by the increasing number of tyrosine kinase inhibitors (TKIs). Here, we characterized the spectrum of 1324 RTK fusions with intact kinase domains in solid tumors by DNA-based high-throughput sequencing. Overall, the prevalence of RTK fusions were 4.7%, with variable frequencies and diverse genomic structures and fusion partners across cancer types. Cancer types, such as thyroid cancers, urological cancers and neuroendocrine tumors are selective in the RTK fusions they carry, while others exhibit highly complex spectra of fusion events. Notably, most RTKs were promiscuous in terms of the partner genes they recombine with. A large proportion of RTK fusions had one of the breakpoints localized to intergenic regions. Comprehensive genomic profiling revealed differences in co-mutational patterns pre- and post-TKI treatments across various RTK fusions. At baseline, multiple cases were detected with co-occurring RTK fusions or concomitant oncogenic mutations in driver genes, such as KRAS and EGFR. Following TKI resistance, we observed differences in potential on- and off-target resistance mutations among fusion variants. For example, the EML4-ALK v3 variant displayed more complex on-target resistance mechanisms, which might explain the reduced survival outcome compared with the v1 variant. Finally, we identified two lung cancer patients with MET+ and NTRK1+ tumors, respectively, who responded well to crizotinib treatment. Taken together, our findings demonstrate the diagnostic and prognostic values of screening for RTK fusions using DNA-based sequencing in solid tumors.
© 2022. The Author(s).
Conflict of interest statement
Jiani C. Yin, Sha Wang, and Yang Shao are employees of Nanjing Geneseeq Technology Inc. All remaining authors have declared no conflict of interests.
Figures



References
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

