An updated definition of V(D)J recombination signal sequences revealed by high-throughput recombination assays
- PMID: 36370096
- PMCID: PMC9723617
- DOI: 10.1093/nar/gkac1038
An updated definition of V(D)J recombination signal sequences revealed by high-throughput recombination assays
Abstract
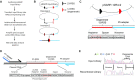
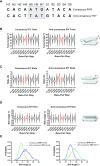
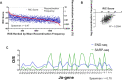
In the adaptive immune system, V(D)J recombination initiates the production of a diverse antigen receptor repertoire in developing B and T cells. Recombination activating proteins, RAG1 and RAG2 (RAG1/2), catalyze V(D)J recombination by cleaving adjacent to recombination signal sequences (RSSs) that flank antigen receptor gene segments. Previous studies defined the consensus RSS as containing conserved heptamer and nonamer sequences separated by a less conserved 12 or 23 base-pair spacer sequence. However, many RSSs deviate from the consensus sequence. Here, we developed a cell-based, massively parallel assay to evaluate V(D)J recombination activity on thousands of RSSs where the 12-RSS heptamer and adjoining spacer region contained randomized sequences. While the consensus heptamer sequence (CACAGTG) was marginally preferred, V(D)J recombination was highly active on a wide range of non-consensus sequences. Select purine/pyrimidine motifs that may accommodate heptamer unwinding in the RAG1/2 active site were generally preferred. In addition, while different coding flanks and nonamer sequences affected recombination efficiency, the relative dependency on the purine/pyrimidine motifs in the RSS heptamer remained unchanged. Our results suggest RAG1/2 specificity for RSS heptamers is primarily dictated by DNA structural features dependent on purine/pyrimidine pattern, and to a lesser extent, RAG:RSS base-specific interactions.
© The Author(s) 2022. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures



References
-
- Schatz D.G., Swanson P.C.. V(D)J recombination: mechanisms of initiation. Annu. Rev. Genet. 2011; 45:167–202. - PubMed
-
- Gellert M. V(D)J recombination: RAG proteins, repair factors, and regulation. Annu. Rev. Biochem. 2002; 71:101–132. - PubMed
-
- Hiom K., Gellert M.. Assembly of a 12/23 paired signal complex: a critical control point in V(D)J recombination. Mol. Cell. 1998; 1:1011–1019. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

