The ProteomeXchange consortium at 10 years: 2023 update
- PMID: 36370099
- PMCID: PMC9825490
- DOI: 10.1093/nar/gkac1040
The ProteomeXchange consortium at 10 years: 2023 update
Abstract
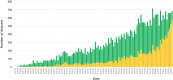
Mass spectrometry (MS) is by far the most used experimental approach in high-throughput proteomics. The ProteomeXchange (PX) consortium of proteomics resources (http://www.proteomexchange.org) was originally set up to standardize data submission and dissemination of public MS proteomics data. It is now 10 years since the initial data workflow was implemented. In this manuscript, we describe the main developments in PX since the previous update manuscript in Nucleic Acids Research was published in 2020. The six members of the Consortium are PRIDE, PeptideAtlas (including PASSEL), MassIVE, jPOST, iProX and Panorama Public. We report the current data submission statistics, showcasing that the number of datasets submitted to PX resources has continued to increase every year. As of June 2022, more than 34 233 datasets had been submitted to PX resources, and from those, 20 062 (58.6%) just in the last three years. We also report the development of the Universal Spectrum Identifiers and the improvements in capturing the experimental metadata annotations. In parallel, we highlight that data re-use activities of public datasets continue to increase, enabling connections between PX resources and other popular bioinformatics resources, novel research and also new data resources. Finally, we summarise the current state-of-the-art in data management practices for sensitive human (clinical) proteomics data.
© The Author(s) 2022. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures

References
-
- Deutsch E.W., Csordas A., Sun Z., Jarnuczak A., Perez-Riverol Y., Ternent T., Campbell D.S., Bernal-Llinares M., Okuda S., Kawano S.et al. .. The proteomexchange consortium in 2017: supporting the cultural change in proteomics public data deposition. Nucleic Acids Res. 2017; 45:D1100–D1106. - PMC - PubMed
-
- Perez-Riverol Y., Bai J., Bandla C., Garcia-Seisdedos D., Hewapathirana S., Kamatchinathan S., Kundu D.J., Prakash A., Frericks-Zipper A., Eisenacher M.et al. .. The PRIDE database resources in 2022: a hub for mass spectrometry-based proteomics evidences. Nucleic Acids Res. 2022; 50:D543–D552. - PMC - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

