Identification of potent compounds against SARs-CoV-2: An in-silico based drug searching against Mpro
- PMID: 36370580
- PMCID: PMC9635257
- DOI: 10.1016/j.compbiomed.2022.106284
Identification of potent compounds against SARs-CoV-2: An in-silico based drug searching against Mpro
Abstract
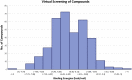
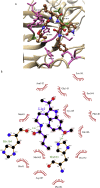
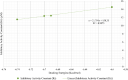
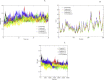
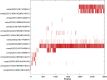
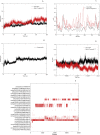
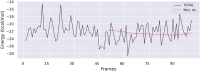
The worldwide pandemic of coronavirus disease 2019 (COVID-19) along with the various newly discovered major SARS-CoV-2 variants, including B.1.1.7, B.1.351, and B.1.1.28, constitute the Variant of Concerns (VOC). It's difficult to keep these variants from spreading over the planet. As a result of these VOCs, the fifth wave has already begun in several countries. The rapid spread of VOCs is posing a serious threat to human civilization. There is currently no specific medicine available for the treatment of COVID-19. Here, we present the findings of methods that used a combination of structure-assisted drug design, virtual screening, and high-throughput screening to swiftly generate lead compounds against Mpro protein of SARs-CoV-2. Therapeutics, in addition to vaccinations, are an essential element of the healthcare response to COVID-19's persistent threat. In the current study, we designed the efficient compounds that may combat all emerging variants of SARs-CoV-2 by targeting the common Mpro protein. The present study was aimed to discover new compounds that may be proposed as new therapeutic agents to treat COVID-19 infection without any adverse effects. For this purpose, a computational-based virtual screening of 352 in-house synthesized compounds library was performed through molecular docking and Molecular Dynamics (MD) simulation approach. As a result, four novel potent compounds were successfully shortlisted by implementing certain pharmacological, physiological, and ADMET criteria i.e., compounds 3, 4, 21, and 22. Furthermore, MD simulations were performed to evaluate the stability and dynamic behavior of these compounds with Mpro complex for about 30 ns. Eventually, compound 22 was found to be highly potent against Mpro protein and was further evaluated by applying 100 ns simulations. Our findings showed that these shortlisted compounds may have potency to treat the COVID-19 infection for which further experimental validation is proposed as part of a follow-up investigation.
Keywords: COVID-19; Molecular docking and simulation; Mpro; Synthetic compounds; Virtual screening.
Copyright © 2022 Elsevier Ltd. All rights reserved.
Conflict of interest statement
Declaration of competing interest The authors declare that there is no conflict of interest.
Figures


Similar articles
-
Targeting SARS-CoV-2 main protease: structure based virtual screening, in silico ADMET studies and molecular dynamics simulation for identification of potential inhibitors.J Biomol Struct Dyn. 2022 May;40(8):3609-3625. doi: 10.1080/07391102.2020.1848636. Epub 2020 Nov 23. J Biomol Struct Dyn. 2022. PMID: 33226303 Free PMC article.
-
Discovery of a "Cocktail" of Potential SARS-COV-2 Main Protease Inhibitors through Virtual Screening of Known Chemical Components of Vitex negundo L. ("Lagundi").Med Chem. 2022;18(3):364-381. doi: 10.2174/1573406417666210618132003. Med Chem. 2022. PMID: 34148541
-
Exploration of Novel Lichen Compounds as Inhibitors of SARS-CoV-2 Mpro: Ligand-Based Design, Molecular Dynamics, and ADMET Analyses.Appl Biochem Biotechnol. 2022 Dec;194(12):6386-6406. doi: 10.1007/s12010-022-04103-3. Epub 2022 Aug 3. Appl Biochem Biotechnol. 2022. PMID: 35921031 Free PMC article.
-
Looking for SARS-CoV-2 Therapeutics Through Computational Approaches.Curr Med Chem. 2023;30(28):3158-3214. doi: 10.2174/0929867329666221004104430. Curr Med Chem. 2023. PMID: 36200217 Review.
-
Computational Approaches to Designing Antiviral Drugs against COVID-19: A Comprehensive Review.Curr Pharm Des. 2023;29(33):2601-2617. doi: 10.2174/0113816128259795231023193419. Curr Pharm Des. 2023. PMID: 37916490 Review.
Cited by
-
Screening compounds for treating the diabetes and COVID-19 from Miao medicine by molecular docking and bioinformatics.Arab J Chem. 2023 Sep;16(9):105001. doi: 10.1016/j.arabjc.2023.105001. Epub 2023 May 18. Arab J Chem. 2023. PMID: 37228247 Free PMC article.
-
Covalent Inhibitors from Saudi Medicinal Plants Target RNA-Dependent RNA Polymerase (RdRp) of SARS-CoV-2.Viruses. 2023 Oct 30;15(11):2175. doi: 10.3390/v15112175. Viruses. 2023. PMID: 38005857 Free PMC article.
-
Exploring the Therapeutic Potential of Petiveria alliacea L. Phytochemicals: A Computational Study on Inhibiting SARS-CoV-2's Main Protease (Mpro).Molecules. 2024 May 27;29(11):2524. doi: 10.3390/molecules29112524. Molecules. 2024. PMID: 38893400 Free PMC article.
References
-
- Organization W.H. vol. 45. June 2021. p. 22. (COVID-19 Weekly Epidemiological Update). 2021.
Publication types
MeSH terms
Substances
Supplementary concepts
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

