The transcription factor IRF2 drives interferon-mediated CD8+ T cell exhaustion to restrict anti-tumor immunity
- PMID: 36370712
- PMCID: PMC9809269
- DOI: 10.1016/j.immuni.2022.10.020
The transcription factor IRF2 drives interferon-mediated CD8+ T cell exhaustion to restrict anti-tumor immunity
Abstract
Type I and II interferons (IFNs) stimulate pro-inflammatory programs that are critical for immune activation, but also induce immune-suppressive feedback circuits that impede control of cancer growth. Here, we sought to determine how these opposing programs are differentially induced. We demonstrated that the transcription factor interferon regulatory factor 2 (IRF2) was expressed by many immune cells in the tumor in response to sustained IFN signaling. CD8+ T cell-specific deletion of IRF2 prevented acquisition of the T cell exhaustion program within the tumor and instead enabled sustained effector functions that promoted long-term tumor control and increased responsiveness to immune checkpoint and adoptive cell therapies. The long-term tumor control by IRF2-deficient CD8+ T cells required continuous integration of both IFN-I and IFN-II signals. Thus, IRF2 is a foundational feedback molecule that redirects IFN signals to suppress T cell responses and represents a potential target to enhance cancer control.
Keywords: CD8(+) T cells; CyTOF; IRF2; T cell exhaustion; adoptive cell transfer; cancer; immunotherapy; interferon gamma; interferon regulatory factor 2; type I interferon.
Copyright © 2022 Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests The authors declare no competing interests.
Figures





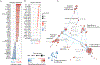
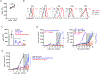
Comment in
-
IRF2 integrates inflammatory signals to balance T cell exhaustion.Immunity. 2022 Dec 13;55(12):2225-2227. doi: 10.1016/j.immuni.2022.11.010. Immunity. 2022. PMID: 36516816
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials

