Modeling community standards for metadata as templates makes data FAIR
- PMID: 36371407
- PMCID: PMC9653497
- DOI: 10.1038/s41597-022-01815-3
Modeling community standards for metadata as templates makes data FAIR
Abstract
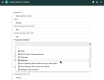
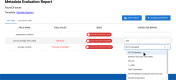
It is challenging to determine whether datasets are findable, accessible, interoperable, and reusable (FAIR) because the FAIR Guiding Principles refer to highly idiosyncratic criteria regarding the metadata used to annotate datasets. Specifically, the FAIR principles require metadata to be "rich" and to adhere to "domain-relevant" community standards. Scientific communities should be able to define their own machine-actionable templates for metadata that encode these "rich," discipline-specific elements. We have explored this template-based approach in the context of two software systems. One system is the CEDAR Workbench, which investigators use to author new metadata. The other is the FAIRware Workbench, which evaluates the metadata of archived datasets for their adherence to community standards. Benefits accrue when templates for metadata become central elements in an ecosystem of tools to manage online datasets-both because the templates serve as a community reference for what constitutes FAIR data, and because they embody that perspective in a form that can be distributed among a variety of software applications to assist with data stewardship and data sharing.
© 2022. The Author(s).
Conflict of interest statement
ES is the Scientific Director of Partners in FAIR (
Figures



References
-
- Bloemers M, Montesanti A. The FAIR funding model: providing a framework for research funders to drive the transition toward FAIR data management and stewardship practices. Data Intelligence. 2020;2(1–2):171–180.
-
- Stall S, et al. Make scientific data FAIR. Nature. 2019;570:27–29. - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources

