Distinct immunological and molecular signatures underpinning influenza vaccine responsiveness in the elderly
- PMID: 36371426
- PMCID: PMC9653450
- DOI: 10.1038/s41467-022-34487-z
Distinct immunological and molecular signatures underpinning influenza vaccine responsiveness in the elderly
Abstract
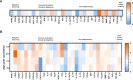
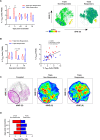
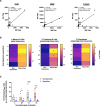
Seasonal influenza outbreaks, especially in high-risk groups such as the elderly, represent an important public health problem. Prevailing inadequate efficacy of seasonal vaccines is a crucial bottleneck. Understanding the immunological and molecular mechanisms underpinning differential influenza vaccine responsiveness is essential to improve vaccination strategies. Here we show comprehensive characterization of the immune response of randomly selected elderly participants (≥ 65 years), immunized with the adjuvanted influenza vaccine Fluad. In-depth analyses by serology, multi-parametric flow cytometry, multiplex and transcriptome analysis, coupled to bioinformatics and mathematical modelling, reveal distinguishing immunological and molecular features between responders and non-responders defined by vaccine-induced seroconversion. Non-responders are specifically characterized by multiple suppressive immune mechanisms. The generated comprehensive high dimensional dataset enables the identification of putative mechanisms and nodes responsible for vaccine non-responsiveness independently of confounding age-related effects, with the potential to facilitate development of tailored vaccination strategies for the elderly.
© 2022. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures






References
-
- (WHO), W.H.O. http://www.who.int/influenza/en/.
-
- de Francisco Shapovalova N, Donadel M, Jit M, Hutubessy R. A systematic review of the social and economic burden of influenza in low- and middle-income countries. Vaccine. 2015;33:6537–6544. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

