Role of the membrane anchor in the regulation of Lck activity
- PMID: 36372231
- PMCID: PMC9763865
- DOI: 10.1016/j.jbc.2022.102663
Role of the membrane anchor in the regulation of Lck activity
Abstract
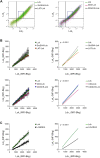
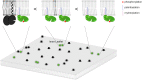
Theoretical work suggests that collective spatiotemporal behavior of integral membrane proteins should be modulated by boundary lipids sheathing their membrane anchors. Here, we show evidence for this prediction while investigating the mechanism for maintaining a steady amount of the active form of integral membrane protein Lck kinase (LckA) by Lck trans-autophosphorylation regulated by the phosphatase CD45. We used super-resolution microscopy, flow cytometry, and pharmacological and genetic perturbation to gain insight into the spatiotemporal context of this process. We found that LckA is generated exclusively at the plasma membrane, where CD45 maintains it in a ceaseless dynamic equilibrium with its unphosphorylated precursor. Steady LckA shows linear dependence, after an initial threshold, over a considerable range of Lck expression levels. This behavior fits a phenomenological model of trans-autophosphorylation that becomes more efficient with increasing LckA. We then challenged steady LckA formation by genetically swapping the Lck membrane anchor with structurally divergent ones, such as that of Src or the transmembrane domains of LAT, CD4, palmitoylation-defective CD4 and CD45 that were expected to drastically modify Lck boundary lipids. We observed small but significant changes in LckA generation, except for the CD45 transmembrane domain that drastically reduced LckA due to its excessive lateral proximity to CD45. Comprehensively, LckA formation and maintenance can be best explained by lipid bilayer critical density fluctuations rather than liquid-ordered phase-separated nanodomains, as previously thought, with "like/unlike" boundary lipids driving dynamical proximity and remoteness of Lck with itself and with CD45.
Keywords: CD45; Lck; boundary lipids; membrane anchor; membrane lateral organization.
Copyright © 2022 The Authors. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Conflict of interest The authors declare no competing interests.
Figures




Similar articles
-
Phosphatase CD45 both positively and negatively regulates T cell receptor phosphorylation in reconstituted membrane protein clusters.J Biol Chem. 2014 Oct 10;289(41):28514-25. doi: 10.1074/jbc.M114.574319. Epub 2014 Aug 15. J Biol Chem. 2014. PMID: 25128530 Free PMC article.
-
Changes in the role of the CD45 protein tyrosine phosphatase in regulating Lck tyrosine phosphorylation during thymic development.J Immunol. 2007 Feb 15;178(4):2056-64. doi: 10.4049/jimmunol.178.4.2056. J Immunol. 2007. PMID: 17277109
-
Tyrosine 192 within the SH2 domain of the Src-protein tyrosine kinase p56Lck regulates T-cell activation independently of Lck/CD45 interactions.Cell Commun Signal. 2020 Nov 23;18(1):183. doi: 10.1186/s12964-020-00673-z. Cell Commun Signal. 2020. PMID: 33225946 Free PMC article.
-
The role of competing mechanisms on Lck regulation.Immunol Res. 2020 Oct;68(5):289-295. doi: 10.1007/s12026-020-09148-2. Epub 2020 Aug 14. Immunol Res. 2020. PMID: 32794043 Review.
-
New insights into the transmembrane protein tyrosine phosphatase CD45.Int J Biochem Cell Biol. 2001 Nov;33(11):1041-6. doi: 10.1016/s1357-2725(01)00075-9. Int J Biochem Cell Biol. 2001. PMID: 11551820 Review.
Cited by
-
Lck Function and Modulation: Immune Cytotoxic Response and Tumor Treatment More Than a Simple Event.Cancers (Basel). 2024 Jul 24;16(15):2630. doi: 10.3390/cancers16152630. Cancers (Basel). 2024. PMID: 39123358 Free PMC article. Review.
-
A non-invasive nanobody probe for high precision mapping of Lck spatial distribution.Front Immunol. 2024 Jul 3;15:1440499. doi: 10.3389/fimmu.2024.1440499. eCollection 2024. Front Immunol. 2024. PMID: 39021567 Free PMC article.
-
T-cell virtuosity in ''knowing thyself".Front Immunol. 2024 Feb 13;15:1343575. doi: 10.3389/fimmu.2024.1343575. eCollection 2024. Front Immunol. 2024. PMID: 38415261 Free PMC article. Review.
-
A Story of Kinases and Adaptors: The Role of Lck, ZAP-70 and LAT in Switch Panel Governing T-Cell Development and Activation.Biology (Basel). 2023 Aug 24;12(9):1163. doi: 10.3390/biology12091163. Biology (Basel). 2023. PMID: 37759563 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

