17q12 deletion syndrome mouse model shows defects in craniofacial, brain and kidney development, and glucose homeostasis
- PMID: 36373506
- PMCID: PMC10655816
- DOI: 10.1242/dmm.049752
17q12 deletion syndrome mouse model shows defects in craniofacial, brain and kidney development, and glucose homeostasis
Abstract
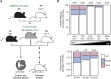
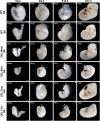
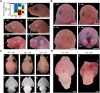
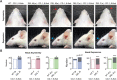
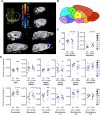
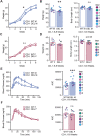
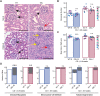
17q12 deletion (17q12Del) syndrome is a copy number variant (CNV) disorder associated with neurodevelopmental disorders and renal cysts and diabetes syndrome (RCAD). Using CRISPR/Cas9 genome editing, we generated a mouse model of 17q12Del syndrome on both inbred (C57BL/6N) and outbred (CD-1) genetic backgrounds. On C57BL/6N, the 17q12Del mice had severe head development defects, potentially mediated by haploinsufficiency of Lhx1, a gene within the interval that controls head development. Phenotypes included brain malformations, particularly disruption of the telencephalon and craniofacial defects. On the CD-1 background, the 17q12Del mice survived to adulthood and showed milder craniofacial and brain abnormalities. We report postnatal brain defects using automated magnetic resonance imaging-based morphometry. In addition, we demonstrate renal and blood glucose abnormalities relevant to RCAD. On both genetic backgrounds, we found sex-specific presentations, with male 17q12Del mice exhibiting higher penetrance and more severe phenotypes. Results from these experiments pinpoint specific developmental defects and pathways that guide clinical studies and a mechanistic understanding of the human 17q12Del syndrome. This mouse mutant represents the first and only experimental model to date for the 17q12 CNV disorder. This article has an associated First Person interview with the first author of the paper.
Keywords: 17q12; Copy number variants; Forebrain development; Head development; Neurodevelopmental disorders; Renal cysts and diabetes syndrome.
© 2022. Published by The Company of Biologists Ltd.
Conflict of interest statement
Competing interests The authors declare no competing or financial interests.
Figures

Similar articles
-
Renal and Extrarenal Phenotypes in Patients With HNF1B Variants and Chromosome 17q12 Microdeletions.Kidney Int Rep. 2024 May 16;9(8):2514-2526. doi: 10.1016/j.ekir.2024.05.007. eCollection 2024 Aug. Kidney Int Rep. 2024. PMID: 39156164 Free PMC article.
-
HNF1β, LHX1, and GGNBP2 deletion contributed to kidney and reproductive dysfunction in 17q12 deletion syndrome: evidence from a case report.Front Genet. 2024 Aug 16;15:1391804. doi: 10.3389/fgene.2024.1391804. eCollection 2024. Front Genet. 2024. PMID: 39221224 Free PMC article.
-
[Clinical and genetic analysis of a fetus with 17q12 microdeletion syndrome].Zhonghua Yi Xue Yi Chuan Xue Za Zhi. 2023 Jun 10;40(6):737-743. doi: 10.3760/cma.j.cn511374-20220828-00583. Zhonghua Yi Xue Yi Chuan Xue Za Zhi. 2023. PMID: 37212013 Chinese.
-
17q12 deletion and duplication syndrome in Denmark-A clinical cohort of 38 patients and review of the literature.Am J Med Genet A. 2016 Nov;170(11):2934-2942. doi: 10.1002/ajmg.a.37848. Epub 2016 Jul 13. Am J Med Genet A. 2016. PMID: 27409573 Review.
-
17q12 Deletion Syndrome as a Rare Cause for Diabetes Mellitus Type MODY5.J Clin Endocrinol Metab. 2018 Oct 1;103(10):3601-3610. doi: 10.1210/jc.2018-00955. J Clin Endocrinol Metab. 2018. PMID: 30032214 Review.
Cited by
-
Prenatal diagnosis and family analysis of 17q12 microdeletion syndrome with fetal renal abnormalities.Front Genet. 2024 Jun 18;15:1401315. doi: 10.3389/fgene.2024.1401315. eCollection 2024. Front Genet. 2024. PMID: 38957807 Free PMC article.
References
-
- Anttonen, A. K., Laari, A., Kousi, M., Yang, Y. J., Jaaskelainen, T., Somer, M., Siintola, E., Jakkula, E., Muona, M., Tegelberg, S.et al. (2017). ZNHIT3 is defective in PEHO syndrome, a severe encephalopathy with cerebellar granule neuron loss. Brain 140, 1267-1279. 10.1093/brain/awx040 - DOI - PubMed
-
- Autism Spectrum Disorders Working Group of The Psychiatric Genomics Consortium. (2017). Meta-analysis of GWAS of over 16,000 individuals with autism spectrum disorder highlights a novel locus at 10q24.32 and a significant overlap with schizophrenia. Mol. Autism 8, 21. 10.1186/s13229-017-0137-9 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Supplementary concepts
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

