Comparative Analysis of Phylogenetic Relationships and Virulence Factor Characteristics between Extended-Spectrum β-Lactamase-Producing Escherichia coli Isolates Derived from Clinical Sites and Chicken Farms
- PMID: 36374015
- PMCID: PMC9769871
- DOI: 10.1128/spectrum.02557-22
Comparative Analysis of Phylogenetic Relationships and Virulence Factor Characteristics between Extended-Spectrum β-Lactamase-Producing Escherichia coli Isolates Derived from Clinical Sites and Chicken Farms
Abstract
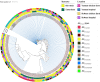
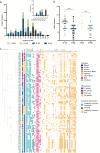
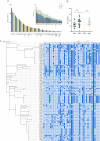
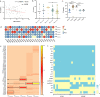
Antimicrobial resistance in bacteria is the most urgent global threat to public health, with extended-spectrum β-lactamase-producing Escherichia coli (ESBL-E. coli) being one of the most documented examples. Nonetheless, the ESBL-E. coli transmission relationship among clinical sites and chicken farms remains unclear. Here, 408 ESBL-E. coli strains were isolated from hospitals and chicken farms in Sichuan Province and Yunnan Province in 2021. We detected blaCTX-M genes in 337 (82.62%) ESBL-E. coli strains. Although the isolation rate, prevalent sequence type (ST) subtypes, and blaCTX-M gene subtypes of ESBL-E. coli varied based on regions and sources, a few strains of CTX-ESBL-E. coli derived from clinical sites and chicken farms in Sichuan Province displayed high genetic similarity. This indicates a risk of ESBL-E. coli transmission from chickens to humans. Moreover, we found that the high-risk clonal strains ST131 and ST1193 primarily carried blaCTX-M-27. This indicates that drug-resistant E. coli from animal and human sources should be monitored. As well, the overuse of β-lactam antibiotics should be avoided in poultry farms to ensure public health and build an effective regulatory mechanism of "farm to fork" under a One Health perspective. IMPORTANCE Bacterial drug resistance has become one of the most significant threats to human health worldwide, especially for extended-spectrum β-lactamase-producing E. coli (ESBL-E. coli). Timely and accurate epidemiological surveys can provide scientific guidance for the adoption of treatments in different regions and also reduce the formation of drug-resistant bacteria. Our study showed that the subtypes of ESBL-E. coli strains prevalent in different provinces are somewhat different, so it is necessary to individualize treatment regimens in different regions, and it is especially important to limit and reduce antibiotic use in poultry farming since chicken-derived ESBL-E. coli serves as an important reservoir of drug resistance genes and has the potential to spread to humans, thus posing a threat to human health. The use of antibiotics in poultry farming should be particularly limited and reduced.
Keywords: ESBL-E. coli; blaCTX-M; chicken farm; hospital; virulence factor.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials

