High-Fidelity Cytosine Base Editing in a GC-Rich Corynebacterium glutamicum with Reduced DNA Off-Target Editing Effects
- PMID: 36374037
- PMCID: PMC9769817
- DOI: 10.1128/spectrum.03760-22
High-Fidelity Cytosine Base Editing in a GC-Rich Corynebacterium glutamicum with Reduced DNA Off-Target Editing Effects
Abstract
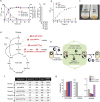
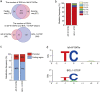
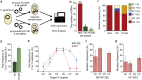
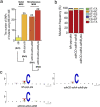
Genome editing technology is a powerful tool for programming microbial cell factories. However, rat APOBEC1-derived cytosine base editor (CBE) that converts C•G to T•A at target genes induced DNA off-targets, regardless of single-guide RNA (sgRNA) sequences. Although the high efficiencies of the bacterial CBEs have been developed, a risk of unidentified off-targets impeded genome editing for microbial cell factories. To address the issues, we demonstrate the genome engineering of Corynebacterium glutamicum as a GC-rich model industrial bacterium by generating premature termination codons (PTCs) in desired genes using high-fidelity cytosine base editors (CBEs). Through this CBE-STOP approach of introducing specific cytosine conversions, we constructed several single-gene-inactivated strains for three genes (ldh, idsA, and pyc) with high base editing efficiencies of average 95.6% (n = 45, C6 position) and the highest success rate of up to 100% for PTCs and ultimately developed a strain with five genes (ldh, actA, ackA, pqo, and pta) that were inactivated sequentially for enhancing succinate production. Although these mutant strains showed the desired phenotypes, whole-genome sequencing (WGS) data revealed that genome-wide point mutations occurred in each strain and further accumulated according to the duration of CBE plasmids. To lower the undesirable mutations, high-fidelity CBEs (pCoryne-YE1-BE3 and pCoryne-BE3-R132E) was employed for single or multiplexed genome editing in C. glutamicum, resulting in drastically reduced sgRNA-independent off-targets. Thus, we provide a CRISPR-assisted bacterial genome engineering tool with an average high efficiency of 90.5% (n = 76, C5 or C6 position) at the desired targets. IMPORTANCE Rat APOBEC1-derived cytosine base editor (CBE) that converts C•G to T•A at target genes induced DNA off-targets, regardless of single-guide RNA (sgRNA) sequences. Although the high efficiencies of bacterial CBEs have been developed, a risk of unidentified off-targets impeded genome editing for microbial cell factories. To address the issues, we identified the DNA off-targets for single and multiple genome engineering of the industrial bacterium Corynebacterium glutamicum using whole-genome sequencing. Further, we developed the high-fidelity (HF)-CBE with significantly reduced off-targets with comparable efficiency and precision. We believe that our DNA off-target analysis and the HF-CBE can promote CRISPR-assisted genome engineering over conventional gene manipulation tools by providing a markerless genetic tool without need for a foreign DNA donor.
Keywords: CRISPR; Corynebacterium glutamicum; cytosine base editor; gene editing; genome editing; industrial bacteria; nonsense mutation; off-target.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


Similar articles
-
Improved plant cytosine base editors with high editing activity, purity, and specificity.Plant Biotechnol J. 2021 Oct;19(10):2052-2068. doi: 10.1111/pbi.13635. Epub 2021 Jun 7. Plant Biotechnol J. 2021. PMID: 34042262 Free PMC article.
-
Transcriptome-wide off-target RNA editing induced by CRISPR-guided DNA base editors.Nature. 2019 May;569(7756):433-437. doi: 10.1038/s41586-019-1161-z. Epub 2019 Apr 17. Nature. 2019. PMID: 30995674 Free PMC article.
-
CRISPR-CBEI: a Designing and Analyzing Tool Kit for Cytosine Base Editor-Mediated Gene Inactivation.mSystems. 2020 Sep 22;5(5):e00350-20. doi: 10.1128/mSystems.00350-20. mSystems. 2020. PMID: 32963098 Free PMC article.
-
Current Status and Challenges of DNA Base Editing Tools.Mol Ther. 2020 Sep 2;28(9):1938-1952. doi: 10.1016/j.ymthe.2020.07.021. Epub 2020 Jul 23. Mol Ther. 2020. PMID: 32763143 Free PMC article. Review.
-
Off-Target Editing by CRISPR-Guided DNA Base Editors.Biochemistry. 2019 Sep 10;58(36):3727-3734. doi: 10.1021/acs.biochem.9b00573. Epub 2019 Aug 26. Biochemistry. 2019. PMID: 31433621 Free PMC article. Review.
Cited by
-
Application progress of CRISPR/Cas9 genome-editing technology in edible fungi.Front Microbiol. 2023 May 25;14:1169884. doi: 10.3389/fmicb.2023.1169884. eCollection 2023. Front Microbiol. 2023. PMID: 37303782 Free PMC article. Review.
-
Genetic tools for metabolic engineering of Pichia pastoris.Eng Microbiol. 2023 Jun 14;3(4):100094. doi: 10.1016/j.engmic.2023.100094. eCollection 2023 Dec. Eng Microbiol. 2023. PMID: 39628915 Free PMC article. Review.
-
Bacterial biosynthesis of abietane-type diterpene ferruginol from glucose.Microb Cell Fact. 2025 Mar 19;24(1):67. doi: 10.1186/s12934-025-02691-3. Microb Cell Fact. 2025. PMID: 40108638 Free PMC article.
-
Tailoring Corynebacterium glutamicum for Sustainable Biomanufacturing: From Traditional to Cutting-Edge Technologies.Mol Biotechnol. 2025 Jun 10. doi: 10.1007/s12033-025-01447-z. Online ahead of print. Mol Biotechnol. 2025. PMID: 40493161 Review.
-
One-for-all gene inactivation via PAM-independent base editing in bacteria.J Biol Chem. 2025 Jan;301(1):108113. doi: 10.1016/j.jbc.2024.108113. Epub 2024 Dec 18. J Biol Chem. 2025. PMID: 39706269 Free PMC article.
References
-
- Luo X, Reiter MA, d’Espaux L, Wong J, Denby CM, Lechner A, Zhang Y, Grzybowski AT, Harth S, Lin W, Lee H, Yu C, Shin J, Deng K, Benites VT, Wang G, Baidoo EEK, Chen Y, Dev I, Petzold CJ, Keasling JD. 2019. Complete biosynthesis of cannabinoids and their unnatural analogues in yeast. Nature 567:123–126. doi:10.1038/s41586-019-0978-9. - DOI - PubMed
-
- Schafer A, Tauch A, Jager W, Kalinowski J, Thierbach G, Puhler A. 1994. Small mobilizable multi-purpose cloning vectors derived from the Escherichia coli plasmids pK18 and pK19: selection of defined deletions in the chromosome of Corynebacterium glutamicum. Gene 145:69–73. doi:10.1016/0378-1119(94)90324-7. - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

