DNA extraction leads to bias in bacterial quantification by qPCR
- PMID: 36374332
- PMCID: PMC10493044
- DOI: 10.1007/s00253-022-12276-4
DNA extraction leads to bias in bacterial quantification by qPCR
Abstract
Quantitative PCR (qPCR) has become a widely used technique for bacterial quantification. The affordability, ease of experimental design, reproducibility, and robustness of qPCR experiments contribute to its success. The establishment of guidelines for minimum information for publication of qPCR experiments, now more than 10 years ago, aimed to mitigate the publication of contradictory data. Unfortunately, there are still a significant number of recent research articles that do not consider the main pitfalls of qPCR for quantification of biological samples, which undoubtedly leads to biased experimental conclusions. qPCR experiments have two main issues that need to be properly tackled: those related to the extraction and purification of genomic DNA and those related to the thermal amplification process. This mini-review provides an updated literature survey that critically analyzes the following key aspects of bacterial quantification by qPCR: (i) the normalization of qPCR results by using exogenous controls, (ii) the construction of adequate calibration curves, and (iii) the determination of qPCR reaction efficiency. It is primarily focused on original papers published last year, where qPCR was applied to quantify bacterial species in different types of biological samples, including multi-species biofilms, human fluids, and water and soil samples. KEY POINTS: • qPCR is a widely used technique used for absolute bacterial quantification. • Recently published papers lack proper qPCR methodologies. • Not including proper qPCR controls significantly affect experimental conclusions.
Keywords: Bacterial load quantification; Calibration curve; Exogenous control; gDNA extraction efficiency; gDNA yield; qPCR; qPCR reaction efficiency.
© 2022. The Author(s), under exclusive licence to Springer-Verlag GmbH Germany, part of Springer Nature.
Conflict of interest statement
Figures
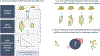
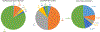


References
-
- Alvarez D, Mendes KF, Tosi M, Fonseca de Souza L, Campos Cedano JC, de Souza Falcão NP, Dunfield K, Tsai SM, Tornisielo VL (2021) Sorption-desorption and biodegradation of sulfometuron-methyl and its effects on the bacterial communities in Amazonian soils amended with aged biochar. Ecotoxicol Environ Saf 207 10.1016/j.ecoenv.2020.111222 - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

