SHERLOCK4HAT: A CRISPR-based tool kit for diagnosis of Human African Trypanosomiasis
- PMID: 36374773
- PMCID: PMC9626900
- DOI: 10.1016/j.ebiom.2022.104308
SHERLOCK4HAT: A CRISPR-based tool kit for diagnosis of Human African Trypanosomiasis
Abstract
Background: To achieve elimination of Human African Trypanosomiasis (HAT) caused by Trypanosoma brucei gambiense (gHAT), the development of highly sensitive diagnostics is needed. We have developed a CRISPR based diagnostic for HAT using SHERLOCK (Specific High-sensitivity Enzymatic Reporter unLOCKing) that is readily adaptable to a field-based setting.
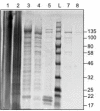
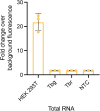
Methods: We adapted SHERLOCK for the detection of T. brucei species. We targeted 7SLRNA, TgSGP and SRA genes and tested SHERLOCK against RNA from blood, buffy coat, dried blood spots (DBS), and clinical samples.
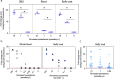
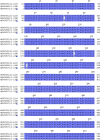
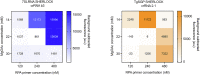
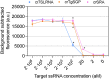
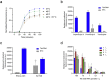
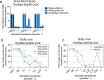
Findings: The pan-Trypanozoon 7SLRNA and T. b. gambiense-specific TgSGP SHERLOCK assays had a sensitivity of 0.1 parasite/μL and a limit of detection 100 molecules/μL. T. b. rhodesiense-specific SRA had a sensitivity of 0.1 parasite/μL and a limit of detection of 10 molecules/μL. TgSGP SHERLOCK and SRA SHERLOCK detected 100% of the field isolated strains. Using clinical specimens from the WHO HAT cryobank, the 7SLRNA SHERLOCK detected trypanosomes in gHAT samples with 56.1%, 95% CI [46.25-65.53] sensitivity and 98.4%, 95% CI [91.41-99.92] specificity, and rHAT samples with 100%, 95% CI [83.18-100] sensitivity and 94.1%, 95% CI [80.91-98.95] specificity. The species-specific TgSGP and SRA SHERLOCK discriminated between the gambiense/rhodesiense HAT infections with 100% accuracy.
Interpretation: The 7SLRNA, TgSGP and SRA SHERLOCK discriminate between gHAT and rHAT infections, and could be used for epidemiological surveillance and diagnosis of HAT in the field after further technical development.
Funding: Institut Pasteur (PTR-175 SHERLOCK4HAT), French Government's Investissement d'Avenir program Laboratoire d'Excellence Integrative Biology of Emerging Infectious Diseases (LabEx IBEID), and Agence Nationale pour la Recherche (ANR-PRC 2021 SherPa).
Keywords: Diagnostic; HAT; SHERLOCK; Trypanosoma brucei.
Copyright © 2022 The Author(s). Published by Elsevier B.V. All rights reserved.
Conflict of interest statement
Declaration of interests The authors declare that they have no competing interests.
Figures









References
-
- Büscher P., Cecchi G., Jamonneau V., Priotto G. Human African trypanosomiasis. Lancet. 2017;390:2397–2409. - PubMed
-
- Büscher P., Gilleman Q., Lejon V. Rapid diagnostic test for sleeping sickness. N Engl J Med. 2013;368:1069–1070. - PubMed
-
- Büscher P., Deborggraeve S. How can molecular diagnostics contribute to the elimination of human African trypanosomiasis? Expert Rev Mol Diagn. 2015;15:607–615. - PubMed
-
- Deborggraeve S., Büscher P. Recent progress in molecular diagnosis of sleeping sickness. Expert Rev Mol Diagn. 2012;12:719–730. - PubMed
MeSH terms
LinkOut - more resources
Full Text Sources

