Targeting platelet GPVI with glenzocimab: a novel mechanism for inhibition
- PMID: 36375047
- PMCID: PMC10119634
- DOI: 10.1182/bloodadvances.2022007863
Targeting platelet GPVI with glenzocimab: a novel mechanism for inhibition
Abstract
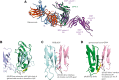
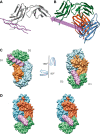
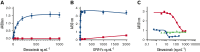
Platelet glycoprotein VI (GPVI) is attracting interest as a potential target for the development of new antiplatelet molecules with a low bleeding risk. GPVI binding to vascular collagen initiates thrombus formation and GPVI interactions with fibrin promote the growth and stability of the thrombus. In this study, we show that glenzocimab, a clinical stage humanized antibody fragment (Fab) with a high affinity for GPVI, blocks the binding of both ligands through a combination of steric hindrance and structural change. A cocrystal of glenzocimab with an extracellular domain of monomeric GPVI was obtained and its structure determined to a resolution of 1.9 Å. The data revealed that (1) glenzocimab binds to the D2 domain of GPVI, GPVI dimerization was not observed in the crystal structure because glenzocimab prevented D2 homotypic interactions and the formation of dimers that have a high affinity for collagen and fibrin; and (2) the light variable domain of the GPVI-bound Fab causes steric hindrance that is predicted to prevent the collagen-related peptide (CRP)/collagen fibers from extending out of their binding site and preclude GPVI clustering and downstream signaling. Glenzocimab did not bind to a truncated GPVI missing loop residues 129 to 136, thus validating the epitope identified in the crystal structure. Overall, these findings demonstrate that the binding of glenzocimab to the D2 domain of GPVI induces steric hindrance and structural modifications that drive the inhibition of GPVI interactions with its major ligands.
© 2023 by The American Society of Hematology. Licensed under Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International (CC BY-NC-ND 4.0), permitting only noncommercial, nonderivative use with attribution. All other rights reserved.
Conflict of interest statement
Conflict-of-interest disclosure: P.B. and M.J.-P. are founders and scientific advisers for Acticor-Biotech. D.F., K.L., and E.T. are employees at Acticor-Biotech. M.W. and N.R. are employees at SARomics Biostructures. The remaining authors declare no competing financial interests.
Figures




References
-
- McFadyen JD, Schaff M, Peter K. Current and future antiplatelet therapies: emphasis on preserving haemostasis. Nat Rev Cardiol. 2018;15(3):181–191. - PubMed
-
- Borst O, Gawaz M. Glycoprotein VI - novel target in antiplatelet medication. Pharmacol Ther. 2021;217:107630. - PubMed
-
- Jandrot-Perrus M, Busfield S, Lagrue AH, et al. Cloning, characterization, and functional studies of human and mouse glycoprotein VI: a platelet-specific collagen receptor from the immunoglobulin superfamily. Blood. 2000;96(5):1798–1807. - PubMed
-
- Nieswandt B, Watson SP. Platelet-collagen interaction: is GPVI the central receptor? Blood. 2003;102(2):449–461. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

