Optimization of deep learning models for the prediction of gene mutations using unsupervised clustering
- PMID: 36376239
- PMCID: PMC9732687
- DOI: 10.1002/cjp2.302
Optimization of deep learning models for the prediction of gene mutations using unsupervised clustering
Abstract
Deep learning models are increasingly being used to interpret whole-slide images (WSIs) in digital pathology and to predict genetic mutations. Currently, it is commonly assumed that tumor regions have most of the predictive power. However, it is reasonable to assume that other tissues from the tumor microenvironment may also provide important predictive information. In this paper, we propose an unsupervised clustering-based multiple-instance deep learning model for the prediction of genetic mutations using WSIs of three cancer types obtained from The Cancer Genome Atlas. Our proposed model facilitates the identification of spatial regions related to specific gene mutations and exclusion of patches that lack predictive information through the use of unsupervised clustering. This results in a more accurate prediction of gene mutations when compared with models using all image patches on WSIs and two recently published algorithms for all three different cancer types evaluated in this study. In addition, our study validates the hypothesis that the prediction of gene mutations solely based on tumor regions on WSI slides may not always provide the best performance. Other tissue types in the tumor microenvironment could provide a better prediction ability than tumor tissues alone. These results highlight the heterogeneity in the tumor microenvironment and the importance of identification of predictive image patches in digital pathology prediction tasks.
Keywords: H&E image; deep learning; gene mutation; unsupervised clustering; whole-slide images.
© 2022 The Authors. The Journal of Pathology: Clinical Research published by The Pathological Society of Great Britain and Ireland and John Wiley & Sons Ltd.
Figures
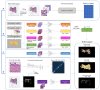
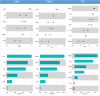


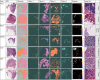

References
-
- Bailey P, Chang DK, Nones K, et al. Genomic analyses identify molecular subtypes of pancreatic cancer. Nature 2016; 531: 47–52. - PubMed
-
- Dienstmann R, Vermeulen L, Guinney J, et al. Consensus molecular subtypes and the evolution of precision medicine in colorectal cancer. Nat Rev Cancer 2017; 17: 79–92. - PubMed
-
- Lindeman NI, Cagle PT, Beasley MB, et al. Molecular testing guideline for selection of lung cancer patients for EGFR and ALK tyrosine kinase inhibitors: guideline from the College of American Pathologists, International Association for the Study of Lung Cancer, and Association for Molecular Pathology. J Thorac Oncol 2013; 8: 823–859. - PMC - PubMed
-
- Russnes HG, Lingjærde OC, Børresen‐Dale A‐L, et al. Breast cancer molecular stratification: from intrinsic subtypes to integrative clusters. Am J Pathol 2017; 187: 2152–2162. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources

