Genome-wide association and multi-trait analyses characterize the common genetic architecture of heart failure
- PMID: 36376295
- PMCID: PMC9663424
- DOI: 10.1038/s41467-022-34216-6
Genome-wide association and multi-trait analyses characterize the common genetic architecture of heart failure
Abstract
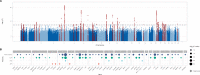
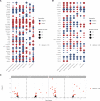
Heart failure is a leading cause of cardiovascular morbidity and mortality. However, the contribution of common genetic variation to heart failure risk has not been fully elucidated, particularly in comparison to other common cardiometabolic traits. We report a multi-ancestry genome-wide association study meta-analysis of all-cause heart failure including up to 115,150 cases and 1,550,331 controls of diverse genetic ancestry, identifying 47 risk loci. We also perform multivariate genome-wide association studies that integrate heart failure with related cardiac magnetic resonance imaging endophenotypes, identifying 61 risk loci. Gene-prioritization analyses including colocalization and transcriptome-wide association studies identify known and previously unreported candidate cardiomyopathy genes and cellular processes, which we validate in gene-expression profiling of failing and healthy human hearts. Colocalization, gene expression profiling, and Mendelian randomization provide convergent evidence for the roles of BCKDHA and circulating branch-chain amino acids in heart failure and cardiac structure. Finally, proteome-wide Mendelian randomization identifies 9 circulating proteins associated with heart failure or quantitative imaging traits. These analyses highlight similarities and differences among heart failure and associated cardiovascular imaging endophenotypes, implicate common genetic variation in the pathogenesis of heart failure, and identify circulating proteins that may represent cardiomyopathy treatment targets.
© 2022. This is a U.S. Government work and not under copyright protection in the US; foreign copyright protection may apply.
Conflict of interest statement
J.D.B. is a full-time employee of Regeneron Genetics Center. E.M.M. consults for Amgen, Avidity, AstraZeneca, Cytokinetics, Janssen, PepGen, Pfizer, Stealth BioTherapeutics, Tenaya Therapeutics, and is a founder of Ikaika Therapeutics. S.M. Damrauer receives research support from RenalytixAI and in-kind research support from Novo Nordisk, as well as personal consulting fees from Calico Labs. The remaining authors declare no competing interests.
Figures





References
-
- Yancy, C. W. et al. 2013 ACCF/AHA guideline for the management of heart failure: a report of the American College of Cardiology Foundation/american Heart Association Task Force on practice guidelines. Circulation128, e240–e327 (2013). - PubMed
-
- Ponikowski P, et al. 2016 ESC Guidelines for the diagnosis and treatment of acute and chronic heart failure: The Task Force for the diagnosis and treatment of acute and chronic heart failure of the European Society of Cardiology (ESC) developed with the special contribution of the Heart Failure Association (HFA) of the ESC. Eur. Heart J. 2016;37:2129–2200. - PubMed
-
- Tsutsui H, et al. JCS 2017/JHFS 2017 Guideline on Diagnosis and Treatment of Acute and Chronic Heart Failure- Digest Version. Circ. J. 2019;83:2084–2184. - PubMed
-
- Bozkurt B, et al. Universal definition and classification of heart failure: a report of the Heart Failure Society of America, Heart Failure Association of the European Society of Cardiology, Japanese Heart Failure Society and Writing Committee of the Universal Definition of Heart Failure. J. Card. Fail. 2021;27:387–413. - PubMed
Publication types
MeSH terms
Grants and funding
- UM1 DK126194/DK/NIDDK NIH HHS/United States
- U01 HG011172/HG/NHGRI NIH HHS/United States
- KL2 TR001879/TR/NCATS NIH HHS/United States
- S10 OD026880/OD/NIH HHS/United States
- R01 DK101478/DK/NIDDK NIH HHS/United States
- UL1 TR001422/TR/NCATS NIH HHS/United States
- I01 CX001737/CX/CSRD VA/United States
- S10 OD030463/OD/NIH HHS/United States
- I01 BX005831/BX/BLRD VA/United States
- P30 DK092949/DK/NIDDK NIH HHS/United States
- TL1 TR001880/TR/NCATS NIH HHS/United States
- R01 HL141232/HL/NHLBI NIH HHS/United States
- R01 HL152446/HL/NHLBI NIH HHS/United States
- R56 DK101478/DK/NIDDK NIH HHS/United States
- I01 BX004821/BX/BLRD VA/United States
- R01 HL105993/HL/NHLBI NIH HHS/United States
- U01 HG011169/HG/NHGRI NIH HHS/United States
- R01 HL128075/HL/NHLBI NIH HHS/United States
- IK2 CX001780/CX/CSRD VA/United States
- T32 HL007843/HL/NHLBI NIH HHS/United States
- F31 AG069441/AG/NIA NIH HHS/United States
LinkOut - more resources
Full Text Sources
Medical

