VIP152 is a selective CDK9 inhibitor with pre-clinical in vitro and in vivo efficacy in chronic lymphocytic leukemia
- PMID: 36376377
- PMCID: PMC9898036
- DOI: 10.1038/s41375-022-01758-z
VIP152 is a selective CDK9 inhibitor with pre-clinical in vitro and in vivo efficacy in chronic lymphocytic leukemia
Abstract
Chronic lymphocytic leukemia (CLL) is effectively treated with targeted therapies including Bruton tyrosine kinase inhibitors and BCL2 antagonists. When these become ineffective, treatment options are limited. Positive transcription elongation factor complex (P-TEFb), a heterodimeric protein complex composed of cyclin dependent kinase 9 (CDK9) and cyclin T1, functions to regulate short half-life transcripts by phosphorylation of RNA Polymerase II (POLII). These transcripts are frequently dysregulated in hematologic malignancies; however, therapies targeting inhibition of P-TEFb have not yet achieved approval for cancer treatment. VIP152 kinome profiling revealed CDK9 as the main enzyme inhibited at 100 nM, with over a 10-fold increase in potency compared with other inhibitors currently in development for this target. VIP152 induced cell death in CLL cell lines and primary patient samples. Transcriptome analysis revealed inhibition of RNA degradation through the AU-Rich Element (ARE) dysregulation. Mechanistically, VIP152 inhibits the assembly of P-TEFb onto the transcription machinery and disturbs binding partners. Finally, immune competent mice engrafted with CLL-like cells of Eµ-MTCP1 over-expressing mice and treated with VIP152 demonstrated reduced disease burden and improvement in overall survival compared to vehicle-treated mice. These data suggest that VIP152 is a highly selective inhibitor of CDK9 that represents an attractive new therapy for CLL.
© 2022. The Author(s).
Conflict of interest statement
AJ, MF, JG, AH, and RI are currently employed by Vincerx Pharma Inc. JG, AH, RI, and JCB are current equity holders in Vincerx Pharma Inc (a publicly traded company) or hold membership on the entity’s Board of Directors or advisory committees. RL is on the scientific advisory board of Vincerx Pharma Inc. JCB holds membership on the Board of Directors or advisory committees of Novartis and Newave and holds Consultancy, Honoraria on Novartis, Trillium Astellas, Astellas, AstraZeneca, Pharmacyclics, and Syndax. J.W. is a consultant for AbbVie Inc, ArQule Inc, AstraZeneca Pharmaceuticals LP, Janssen Biotech Inc, Pharmacyclics LLC, an AbbView Company, is on the advisory committee of AbbVie Inc, ArQule Inc, Janssen Biotech Inc, AstraZeneca, and Beigene. JW has received research funding from AbbVie Inc, and Loxo Oncology Inc. JB holds consultancy and honoraria from AbbVie Inc. AstraZeneca, Kite, and Innate. All other authors declare no conflicts of interest.
Figures
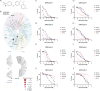



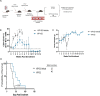

References
-
- Cheson BD, Meyer RM. Chronic lymphocytic leukemia. N. Engl J Med. 2005;352:804–15. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

