The protein conformational basis of isoflavone biosynthesis
- PMID: 36376429
- PMCID: PMC9663428
- DOI: 10.1038/s42003-022-04222-x
The protein conformational basis of isoflavone biosynthesis
Abstract
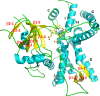
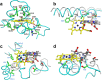
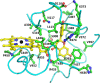
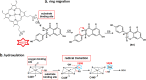
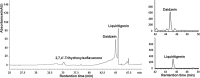
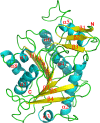
Isoflavonoids play important roles in plant defense and also exhibit a range of mammalian health-promoting activities. Their biosynthesis is initiated by two enzymes with unusual catalytic activities; 2-hydroxyisoflavanone synthase (2-HIS), a membrane-bound cytochrome P450 catalyzing a coupled aryl-ring migration and hydroxylation, and 2-hydroxyisoflavanone dehydratase (2-HID), a member of a large carboxylesterase family that paradoxically catalyzes dehydration of 2-hydroxyisoflavanones to isoflavone. Here we report the crystal structures of 2-HIS from Medicago truncatula and 2-HID from Pueraria lobata. The 2-HIS structure reveals a unique cytochrome P450 conformation and heme and substrate binding mode that facilitate the coupled aryl-ring migration and hydroxylation reactions. The 2-HID structure reveals the active site architecture and putative catalytic residues for the dual dehydratase and carboxylesterase activities. Mutagenesis studies revealed key residues involved in substrate binding and specificity. Understanding the structural basis of isoflavone biosynthesis will facilitate the engineering of new bioactive isoflavonoids.
© 2022. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures
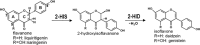
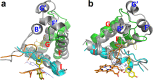
Similar articles
-
Molecular and biochemical characterization of 2-hydroxyisoflavanone dehydratase. Involvement of carboxylesterase-like proteins in leguminous isoflavone biosynthesis.Plant Physiol. 2005 Mar;137(3):882-91. doi: 10.1104/pp.104.056747. Epub 2005 Feb 25. Plant Physiol. 2005. PMID: 15734910 Free PMC article.
-
2-Hydroxyisoflavanone dehydratase is a critical determinant of isoflavone productivity in hairy root cultures of Lotus japonicus.Plant Cell Physiol. 2007 Nov;48(11):1652-7. doi: 10.1093/pcp/pcm125. Epub 2007 Oct 4. Plant Cell Physiol. 2007. PMID: 17921150
-
Key amino acid residues required for aryl migration catalysed by the cytochrome P450 2-hydroxyisoflavanone synthase.Plant J. 2002 Sep;31(5):555-64. doi: 10.1046/j.1365-313x.2002.01378.x. Plant J. 2002. PMID: 12207646
-
Differential behavior of the sub-sites of cytochrome 450 active site in binding of substrates, and products (implications for coupling/uncoupling).Biochim Biophys Acta. 2007 Mar;1770(3):360-75. doi: 10.1016/j.bbagen.2006.09.018. Epub 2006 Oct 5. Biochim Biophys Acta. 2007. PMID: 17134838 Review.
-
Mechanism and role of covalent heme binding in the CYP4 family of P450 enzymes and the mammalian peroxidases.Drug Metab Rev. 2008;40(3):405-26. doi: 10.1080/03602530802186439. Drug Metab Rev. 2008. PMID: 18642140 Review.
Cited by
-
Key genes in a "Galloylation-Degalloylation cycle" controlling the synthesis of hydrolyzable tannins in strawberry plants.Hortic Res. 2024 Dec 16;12(4):uhae350. doi: 10.1093/hr/uhae350. eCollection 2025 Apr. Hortic Res. 2024. PMID: 40046039 Free PMC article.
-
Evolution and diversification of carboxylesterase-like [4+2] cyclases in aspidosperma and iboga alkaloid biosynthesis.Proc Natl Acad Sci U S A. 2024 Feb 13;121(7):e2318586121. doi: 10.1073/pnas.2318586121. Epub 2024 Feb 6. Proc Natl Acad Sci U S A. 2024. PMID: 38319969 Free PMC article.
-
Advances in synthesizing plant-derived isoflavones and their precursors with multiple pharmacological activities using engineered yeasts.Microb Cell Fact. 2025 Mar 29;24(1):75. doi: 10.1186/s12934-025-02692-2. Microb Cell Fact. 2025. PMID: 40155940 Free PMC article. Review.
-
Systematic Engineering of Saccharomyces cerevisiae for the De Novo Biosynthesis of Genistein and Glycosylation Derivatives.J Fungi (Basel). 2024 Feb 26;10(3):176. doi: 10.3390/jof10030176. J Fungi (Basel). 2024. PMID: 38535185 Free PMC article.
-
Discrepancy of Growth Toxicity of Polystyrene Nanoplastics on Soybean (Glycine max) and Mung Bean (Vigna radiata).Toxics. 2024 Feb 17;12(2):155. doi: 10.3390/toxics12020155. Toxics. 2024. PMID: 38393250 Free PMC article.
References
-
- Dixon, R. A. Isoflavonoids: biochemistry, molecular biology and biological functions. in Comprehensive Natural Products Chemistry (ed. Sankawa, U.) 773–823 (Elsevier, 1999).
-
- Dixon RA, Ferreira D. Molecules of interest: genistein. Phytochemistry. 2002;60:205–211. - PubMed
-
- Palevitz BA. Soybeans hit main street. Scientist. 2000;14:8–9.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

