A novel bi-directional heterogeneous network selection method for disease and microbial association prediction
- PMID: 36376802
- PMCID: PMC9664813
- DOI: 10.1186/s12859-022-04961-y
A novel bi-directional heterogeneous network selection method for disease and microbial association prediction
Abstract
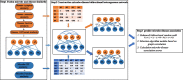
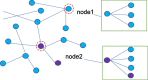
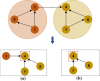
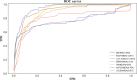
Microorganisms in the human body have a great impact on human health. Therefore, mastering the potential relationship between microorganisms and diseases is helpful to understand the pathogenesis of diseases and is of great significance to the prevention, diagnosis, and treatment of diseases. In order to predict the potential microbial disease relationship, we propose a new computational model. Firstly, a bi-directional heterogeneous microbial disease network is constructed by integrating multiple similarities, including Gaussian kernel similarity, microbial function similarity, disease semantic similarity, and disease symptom similarity. Secondly, the neighbor information of the network is learned by random walk; Finally, the selection model is used for information aggregation, and the microbial disease node pair is analyzed. Our method is superior to the existing methods in leave-one-out cross-validation and five-fold cross-validation. Moreover, in case studies of different diseases, our method was proven to be effective.
Keywords: Bi-directional heterogeneous network; Potential microorganism disease prediction; Random walk; causal selection model.
© 2022. The Author(s).
Conflict of interest statement
The authors declare that they have no competing interests.
Figures
References
-
- Chen X, Huang YA, You ZH, et al. A novel approach based on KATZ measure to predict associations of human microbiota with non-infectious diseases. Bioinformatics. 2017;33:733–739. - PubMed
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

