Meta-analysis of 16S rRNA microbial data identified alterations of the gut microbiota in COVID-19 patients during the acute and recovery phases
- PMID: 36376804
- PMCID: PMC9662111
- DOI: 10.1186/s12866-022-02686-9
Meta-analysis of 16S rRNA microbial data identified alterations of the gut microbiota in COVID-19 patients during the acute and recovery phases
Abstract
Background: Dozens of studies have demonstrated gut dysbiosis in COVID-19 patients during the acute and recovery phases. However, a consensus on the specific COVID-19 associated bacteria is missing. In this study, we performed a meta-analysis to explore whether robust and reproducible alterations in the gut microbiota of COVID-19 patients exist across different populations.
Methods: A systematic review was conducted for studies published prior to May 2022 in electronic databases. After review, we included 16 studies that comparing the gut microbiota in COVID-19 patients to those of controls. The 16S rRNA sequence data of these studies were then re-analyzed using a standardized workflow and synthesized by meta-analysis.
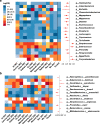
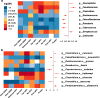
Results: We found that gut bacterial diversity of COVID-19 patients in both the acute and recovery phases was consistently lower than non-COVID-19 individuals. Microbial differential abundance analysis showed depletion of anti-inflammatory butyrate-producing bacteria and enrichment of taxa with pro-inflammatory properties in COVID-19 patients during the acute phase compared to non-COVID-19 individuals. Analysis of microbial communities showed that the gut microbiota of COVID-19 recovered patients were still in unhealthy ecostates.
Conclusions: Our results provided a comprehensive synthesis to better understand gut microbial perturbations associated with COVID-19 and identified underlying biomarkers for microbiome-based diagnostics and therapeutics.
Keywords: 16S rRNA; Acute phase; COVID-19; Gut microbiota; Recovery phase.
© 2022. The Author(s).
Conflict of interest statement
The authors declare that they have no competing interests.
Figures


Similar articles
-
Gut microbiota alterations in colorectal adenoma-carcinoma sequence based on 16S rRNA gene sequencing: A systematic review and meta-analysis.Microb Pathog. 2024 Oct;195:106889. doi: 10.1016/j.micpath.2024.106889. Epub 2024 Aug 26. Microb Pathog. 2024. PMID: 39197689
-
Correlation Analysis between Gut Microbiota Alterations and the Cytokine Response in Patients with Coronavirus Disease during Hospitalization.Microbiol Spectr. 2022 Apr 27;10(2):e0168921. doi: 10.1128/spectrum.01689-21. Epub 2022 Mar 7. Microbiol Spectr. 2022. PMID: 35254122 Free PMC article.
-
Uncovering specific taxonomic and functional alteration of gut microbiota in chronic kidney disease through 16S rRNA data.Front Cell Infect Microbiol. 2024 Apr 19;14:1363276. doi: 10.3389/fcimb.2024.1363276. eCollection 2024. Front Cell Infect Microbiol. 2024. PMID: 38707511 Free PMC article.
-
Gut Microbiome Alterations and Functional Prediction in Chronic Spontaneous Urticaria Patients.J Microbiol Biotechnol. 2021 May 28;31(5):747-755. doi: 10.4014/jmb.2012.12022. J Microbiol Biotechnol. 2021. PMID: 33746191 Free PMC article.
-
Alterations of Gut Microbiota in Patients With Irritable Bowel Syndrome Based on 16S rRNA-Targeted Sequencing: A Systematic Review.Clin Transl Gastroenterol. 2019 Feb;10(2):e00012. doi: 10.14309/ctg.0000000000000012. Clin Transl Gastroenterol. 2019. PMID: 30829919 Free PMC article.
Cited by
-
The Multisystem Impact of Long COVID: A Comprehensive Review.Diagnostics (Basel). 2024 Jan 24;14(3):244. doi: 10.3390/diagnostics14030244. Diagnostics (Basel). 2024. PMID: 38337760 Free PMC article. Review.
-
Infection, Dysbiosis and Inflammation Interplay in the COVID Era in Children.Int J Mol Sci. 2023 Jun 29;24(13):10874. doi: 10.3390/ijms241310874. Int J Mol Sci. 2023. PMID: 37446047 Free PMC article. Review.
-
The microbiome in post-acute infection syndrome (PAIS).Comput Struct Biotechnol J. 2023 Aug 5;21:3904-3911. doi: 10.1016/j.csbj.2023.08.002. eCollection 2023. Comput Struct Biotechnol J. 2023. PMID: 37602232 Free PMC article. Review.
-
Gut Microbiome Disruption Following SARS-CoV-2: A Review.Microorganisms. 2024 Jan 9;12(1):131. doi: 10.3390/microorganisms12010131. Microorganisms. 2024. PMID: 38257958 Free PMC article. Review.
-
COVID-19 and Gastrointestinal Tract: From Pathophysiology to Clinical Manifestations.Medicina (Kaunas). 2023 Sep 24;59(10):1709. doi: 10.3390/medicina59101709. Medicina (Kaunas). 2023. PMID: 37893427 Free PMC article. Review.
References
-
- Aleem A, Akbar Samad AB, Slenker AK. Emerging Variants of SARS-CoV-2 And Novel Therapeutics Against Coronavirus (COVID-19). Treasure Island (FL); 2022. - PubMed
-
- Li J, Lai S, Gao GF, Shi W. The emergence, genomic diversity and global spread of SARS-CoV-2. Nature. 2021;600(7889):408–418. - PubMed
-
- Sencio V, Barthelemy A, Tavares LP, Machado MG, Soulard D, Cuinat C, et al. Gut Dysbiosis during influenza contributes to pulmonary pneumococcal Superinfection through altered short-Chain fatty acid production. Cell Rep. 2020;30(9):2934–2947.e6. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

