Ecological Speciation Promoted by Divergent Regulation of Functional Genes Within African Cichlid Fishes
- PMID: 36376993
- PMCID: PMC10101686
- DOI: 10.1093/molbev/msac251
Ecological Speciation Promoted by Divergent Regulation of Functional Genes Within African Cichlid Fishes
Abstract
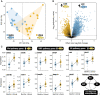
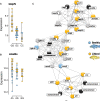
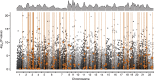
Rapid ecological speciation along depth gradients has taken place repeatedly in freshwater fishes, yet molecular mechanisms facilitating such diversification are typically unclear. In Lake Masoko, an African crater lake, the cichlid Astatotilapia calliptera has diverged into shallow-littoral and deep-benthic ecomorphs with strikingly different jaw structures within the last 1,000 years. Using genome-wide transcriptome data, we explore two major regulatory transcriptional mechanisms, expression and splicing-QTL variants, and examine their contributions to differential gene expression underpinning functional phenotypes. We identified 7,550 genes with significant differential expression between ecomorphs, of which 5.4% were regulated by cis-regulatory expression QTLs, and 9.2% were regulated by cis-regulatory splicing QTLs. We also found strong signals of divergent selection on differentially expressed genes associated with craniofacial development. These results suggest that large-scale transcriptome modification plays an important role during early-stage speciation. We conclude that regulatory variants are important targets of selection driving ecologically relevant divergence in gene expression during adaptive diversification.
Keywords: cichlids; ecological speciation; gene expression; molecular evolution; transcriptional regulation.
© The Author(s) 2022. Published by Oxford University Press on behalf of Society for Molecular Biology and Evolution.
Figures


References
-
- Abzhanov A, Kuo WP, Hartmann C, Grant BR, Grant PR, Tabin CJ. 2006. The calmodulin pathway and evolution of elongated beak morphology in Darwin's Finches. Nature. 442:563–567. - PubMed
-
- Abzhanov A, Protas M, Grant BR, Grant PR, Tabin CJ. 2004. Bmp4 and morphological variation of beaks in Darwin's Finches. Science. 305:1462–1465. - PubMed
-
- Albertson RC. 2008. Morphological divergence predicts habitat partitioning in a Lake Malawi cichlid species complex. Copeia. 2008:689–698.
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

