High-throughput morphometric and transcriptomic profiling uncovers composition of naïve and sensory-deprived cortical cholinergic VIP/CHAT neurons
- PMID: 36377476
- PMCID: PMC9811618
- DOI: 10.15252/embj.2021110565
High-throughput morphometric and transcriptomic profiling uncovers composition of naïve and sensory-deprived cortical cholinergic VIP/CHAT neurons
Abstract
Cortical neuronal networks control cognitive output, but their composition and modulation remain elusive. Here, we studied the morphological and transcriptional diversity of cortical cholinergic VIP/ChAT interneurons (VChIs), a sparse population with a largely unknown function. We focused on VChIs from the whole barrel cortex and developed a high-throughput automated reconstruction framework, termed PopRec, to characterize hundreds of VChIs from each mouse in an unbiased manner, while preserving 3D cortical coordinates in multiple cleared mouse brains, accumulating thousands of cells. We identified two fundamentally distinct morphological types of VChIs, bipolar and multipolar that differ in their cortical distribution and general morphological features. Following mild unilateral whisker deprivation on postnatal day seven, we found after three weeks both ipsi- and contralateral dendritic arborization differences and modified cortical depth and distribution patterns in the barrel fields alone. To seek the transcriptomic drivers, we developed NuNeX, a method for isolating nuclei from fixed tissues, to explore sorted VChIs. This highlighted differentially expressed neuronal structural transcripts, altered exitatory innervation pathways and established Elmo1 as a key regulator of morphology following deprivation.
Keywords: 3D; ChAT; deprivation; interneurons; reconstruction.
© 2022 The Authors. Published under the terms of the CC BY NC ND 4.0 license.
Figures

ChAT‐CreXAi14 mice were sacrificed, perfused, and cortices were isolated and subjected to iDISCO+ clearing and light‐sheet imaging.
PopRec pipeline preprocessing: Across‐sample signal intensity correction with the intensify3D+ App. Intensity gradient correction: Precorrection (red frame); postcorrection (blue frame); Intensity profiles correspond to yellow horizontal lines. Across‐sample intensity was corrected by mean pixel histogram equilibration.
PopRec pipeline step 1: automatic soma detection and data augmentation into high‐resolution mini‐cubes around each cell together with neighboring cells and dendrites with the Soma‐to‐Cube App.
PopRec pipeline step 2: feeding every mini‐cube to a computer cluster node together with a combination of two reconstruction parameters (maximal point gap distance (MPD) and fluorescence threshold (Thr)) with the Neuron reconstruction App (Architecture based on Trees Toolbox competitive reconstruction) while accounting for close proximity somata.
PopRec pipeline step 3: feature extraction (BlueBrain/NeuroM) and ranking of candidate reconstruction in a randomized manner with the Reconstruction validation App where the user decides if the current reconstruction displayed is accurate or not with full 3D visualization of original imaging.
The output of the PopRec pipeline produces the reconstruction cubes, neuronal reconstructions, a list of features from the reconstructed neurons, and the predicted rank of each neuron in the training dataset. In this study, we used this subset rank to train a regression model (MATLAB) and filter out neurons that do not pass a certain quality threshold from downstream analysis.
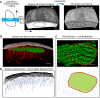
Scheme of light‐sheet autofluorescence imaging of the barrel cortex region and Intensify3D+ normalization of autofluorescence pixel histograms, which corrects 3D light‐sheet scanning artifacts (white arrows) and variable intensity (black ellipse) in a tissue volume. Scale bars: 0.5 mm.
Volume rendering of images postnormalization used to isolate the cortical surface and estimate the boundaries of barrel regions (green) without any additional staining, large blood vessels are depicted in red. Scale bars: 0.5 mm.
A 3D barrel cortex region of interest (ROI) area (top—red line) is manually measured and segmented (FIJI) to define an anatomically based ROI (bottom). This ROI is used to determine for all neuronal somata (blue dots) whether they are located within or outside of the barrel cortex. Scale bars: 0.5 mm.
3D registration and mesh segmentation of VChI somata to the cortical surface. The minimal Euclidean distance and direction (blue arrows) to the surface are calculated for each of the neuron somata (red dots).

Sample collection: naïve mouse cortices were processed, the barrel cortex area was defined, and VChIs were subjected to the PopRec pipeline.
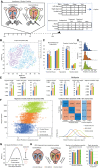
tSNE clustering by morphological features. Colors represent the number of stem dendrites or dendritic trees originating from the cell soma.
Representative bipolar (left) and multipolar (right) VChIs of each group. Horizontal scale bar: 100 μm.
Cortical depth distribution of biVChIs (blue) and mVChIs (green) measured automatically by soma detection.
3D Sholl analysis of biVChIs and mVChIs (Bonferroni corrected linear mixed effects, LME, error bars represent SEM+‐). Example of Sholl analysis and number of intersections (right). Bars represent the standard error of the mean.
Distribution of biVChIs vs mVChIs for total neuron length, mean stem dendrite length per neuron, mean number of bifurcations per neuron, stem dendrite (SD) compression as a measure of dendrite curvature, and the total number of sections per neuron. See Dataset EV4 for full statistical description.
PatchSeq analysis of VChIs. UMAP embedding of expression profiles from the entire study highlighting marker genes for inhibitory interneurons. Bottom right—only ChAT‐positive neurons out of which 14 VChIs from layer 2/3 (n = 8 bipolar and n = 6 multipolar) labeled as bipolar and multipolar.
Experimental scheme: mice were subjected to a single event of anesthesia with or without unilateral whisker plucking at p7. Mouse brains were collected at p30 and subjected to the PopRec pipeline.
tSNE clustering of whisker‐deprived mice and controls.
Observed shift in cell type proportion and depth distribution in both ipsi‐ and contralateral hemispheres (cross tab test with post hoc).
Cell depth distributions from all three experimental conditions; notice the increased proportion of deep VChIs in whisker‐deprived mice. Dashed line marks 0.38 relative depth (surface to callosum).
Specific features of mVChIs and biVChIs to whisker deprivation (Linear mixed effects, error bars represent SEM+‐).
Regression model for predicting cortical condition at the single‐cell level. Left—prediction value for each cell. Top right—confusion matrix for classification of neurons to condition chance level is 33%. Bottom right—prediction summary histogram.
Ripley's L statistics for spatial distribution in 3D as a function of relative distance.
Peripheral barrel selection and proportion of bipolar and multipolar VChIs as well as depth distribution of peripheral VChIs (Dataset EV10, Cross tab test with post hoc).
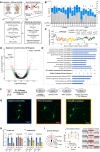
Experimental design and NuNeX RNA isolation strategy. Mice underwent unilateral whisker deprivation on day p7, were sacrificed on day 30 and their barrel cortices were isolated, lightly fixed, homogenized, and subjected to fluorescence‐activated cell sorting (FACS). RNA was extracted from pool‐sorted tdTomato and DAPI‐positive neurons from left and right cortices of different mice, subjected to RNA‐seq and downstream analysis.
RNA‐sequencing and mapping statistics.
Correlation analysis of Cholinergic and VIP‐related transcript profiles from control mice to a web‐available dataset (Allen Institute) of 246 neuron types. ChAT/VIP neurons are most tightly correlated with bulk RNA profiles from control mice.
Volcano plot depicting 110 DE transcripts between un‐deprived (n = 4 pools of 1–3 mice, 8 samples – 4L, 4R) and deprived mice (n = 6 pools of 1–3 mice, 12 samples – 6L, 6R, DeSeq2 differential expression model).
Pathway enrichment analysis of cellular compartments altered between control and deprived mice.
siPools KD of Arpp21, Sam5b and Elmo1 transcripts in cultured primary neurons.
Example images of Tuj1 labeled primary neurons under various KD conditions.
Bar plots represent the expression of Elmo1 and Sema5b transcripts following KD of either transcript.
Sholl analysis of primary neurons at 20–80 micron radius from soma center (ANOVA 1 with Tukey post hoc error bars represent SEM).
References
-
- Altman RB, Keith Dunker A, Hunter L, Ritchie MD, Murray TA, Klein TE (2016) Biocomputing 2017: proceedings of the Pacific symposium. Singapore: World Scientific Publishing Company;
-
- Antonopoulos CG, Rubido N, Batista A, Baptista MS (2021) Advancing our understanding of structure and function in the brain: developing novel approaches for network inference and emergent phenomena. Lausanne: Frontiers Media SA;
-
- Beniaguev D, Segev I, London M (2021) Single cortical neurons as deep artificial neural networks. Neuron 109: 2727–2739 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

