Molecular basis of astrocyte diversity and morphology across the CNS in health and disease
- PMID: 36378959
- PMCID: PMC9873482
- DOI: 10.1126/science.adc9020
Molecular basis of astrocyte diversity and morphology across the CNS in health and disease
Abstract
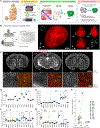
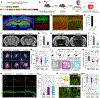
Astrocytes, a type of glia, are abundant and morphologically complex cells. Here, we report astrocyte molecular profiles, diversity, and morphology across the mouse central nervous system (CNS). We identified shared and region-specific astrocytic genes and functions and explored the cellular origins of their regional diversity. We identified gene networks correlated with astrocyte morphology, several of which unexpectedly contained Alzheimer's disease (AD) risk genes. CRISPR/Cas9-mediated reduction of candidate genes reduced astrocyte morphological complexity and resulted in cognitive deficits. The same genes were down-regulated in human AD, in an AD mouse model that displayed reduced astrocyte morphology, and in other human brain disorders. We thus provide comprehensive molecular data on astrocyte diversity and mechanisms across the CNS and on the molecular basis of astrocyte morphology in health and disease.
Figures




Comment in
-
Molecular diversity of astrocytes.Science. 2022 Nov 4;378(6619):475-476. doi: 10.1126/science.ade9249. Epub 2022 Nov 3. Science. 2022. PMID: 36378974
References
-
- Bargmann C et al., BRAIN 2025: a scientific vision. Brain Research through Advancing Innovative Neurotechnologies (BRAIN) Working Group Report to the Advisory Committee to the Director, NIH, (2014).
-
- Barres BA, The mystery and magic of glia: a perspective on their roles in health and disease. Neuron 60, 430–440 (2008). - PubMed
-
- Yu X, Nagai J, Khakh BS, Improved tools to study astrocytes. Nature reviews. Neuroscience 21, 121–138 (2020). - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials

