Genome-wide profiling of histone modifications in Plasmodium falciparum using CUT&RUN
- PMID: 36379668
- PMCID: PMC9670794
- DOI: 10.26508/lsa.202201778
Genome-wide profiling of histone modifications in Plasmodium falciparum using CUT&RUN
Abstract
We recently adapted a CUT&RUN protocol for genome-wide profiling of chromatin modifications in the human malaria parasite Plasmodium Using the step-by-step protocol described below, we were able to generate high-quality profiles of multiple histone modifications using only a small fraction of the cells required for ChIP-seq. Using antibodies against two commonly profiled histone modifications, H3K4me3 and H3K9me3, we show here that CUT&RUN profiling is highly reproducible and closely recapitulates previously published ChIP-seq-based abundance profiles of histone marks. Finally, we show that CUT&RUN requires substantially lower sequencing coverage for accurate profiling compared with ChIP-seq.
© 2022 Morillo et al.
Conflict of interest statement
The authors declare that they have no conflicts of interest.
Figures

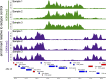



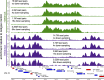



References
-
- Bártfai R, Hoeijmakers WA, Salcedo-Amaya AM, Smits AH, Janssen-Megens E, Kaan A, Treeck M, Gilberger TW, Françoijs KJ, Stunnenberg HG (2010) H2A.Z demarcates intergenic regions of the plasmodium falciparum epigenome that are dynamically marked by H3K9ac and H3K4me3. PLoS Pathog 6: e1001223. 10.1371/journal.ppat.1001223 - DOI - PMC - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
