IRF1 Is Required for MDA5 (IFIH1) Induction by IFN-α, LPS, and poly(I:C) in Murine Macrophages
- PMID: 36380629
- PMCID: PMC10643899
- DOI: 10.1159/000527008
IRF1 Is Required for MDA5 (IFIH1) Induction by IFN-α, LPS, and poly(I:C) in Murine Macrophages
Erratum in
-
Erratum.J Innate Immun. 2025;17(1):287. doi: 10.1159/000545776. Epub 2025 Jun 11. J Innate Immun. 2025. PMID: 40499528 Free PMC article. No abstract available.
Abstract
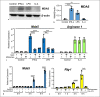
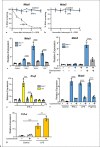
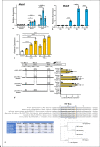
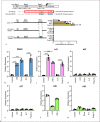
Melanoma differentiation-associated protein 5 (MDA5) induces type I interferons (IFNs) after the recognition of viral RNA. In addition, gain-of-function mutations in the interferon induced with helicase C domain 1 (IFIH1) gene, which encodes MDA5, lead to type I interferonopathies. Here, we show that Mda5 is highly expressed in murine macrophages and is regulated by pro-inflammatory stimuli such as the cytokines IFN-α and IFN-γ, the TLR ligand LPS, and a mimic of dsRNA, poly(I:C). Mda5 induction is mediated through the production of reactive oxygen species. The induction by IFN-α or LPS occurs at the transcriptional level since the Mda5 mRNA half-life before and after induction is very stable. Interestingly, STAT1 is required for Mda5 induction by IFN-α, LPS, or poly(I:C). The time course of induction of at least 3 h and the need for protein synthesis indicate that Mda5 requires an intermediate protein for transcription. In transient transfection experiments, we found that a 105-bp fragment of this gene, between -1153 and -1258 bp relative to the transcription start site, is required for transcription. In this specific region, we observed a sequence containing an IRF-binding motif, which, when mutated, abolishes the induction of Mda5. This sequence is strongly conserved in the IFIH1 promoters of eutherian mammals and in other distant species. Kinetic experiments, chromatin immunoprecipitation assays, and gene-silencing experiments revealed that IRF1 is required for induction of Mda5 expression.
Keywords: Gene regulation; Inflammation; Interferons; Macrophages; Reactive oxygen species.
© 2022 The Author(s). Published by S. Karger AG, Basel.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures


References
-
- Honda K, Takaoka A, Taniguchi T. Type I interferon [corrected] gene induction by the interferon regulatory factor family of transcription factors. Immunity. 2006 Sep;25((3)):349–360. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

