Biomimetic nanoparticles drive the mechanism understanding of shear-wave elasticity stiffness in triple negative breast cancers to predict clinical treatment
- PMID: 36382024
- PMCID: PMC9638718
- DOI: 10.1016/j.bioactmat.2022.10.025
Biomimetic nanoparticles drive the mechanism understanding of shear-wave elasticity stiffness in triple negative breast cancers to predict clinical treatment
Abstract
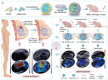
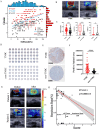
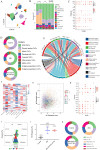
In clinical practice, we noticed that triple negative breast cancer (TNBC) patients had higher shear-wave elasticity (SWE) stiffness than non-TNBC patients and a higher α-SMA expression was found in TNBC tissues than the non-TNBC tissues. Moreover, SWE stiffness also shows a clear correlation to neoadjuvant response efficiency. To elaborate this phenomenon, TNBC cell membrane-modified polylactide acid-glycolic acid (PLGA) nanoparticle was fabricated to specifically deliver artesunate to regulate SWE stiffness through inhibiting CAFs functional status. As tested in MDA-MB-231 and E0771 orthotopic tumor models, CAFs functional status inhibited by 231M-ARS@PLGA nanoparticles (231M-AP NPs) had reduced the SWE stiffness as well as attenuated hypoxia of tumor as tumor soil loosening agent which amplified the antitumor effects of paclitaxel and PD1 inhibitor. Single-cell sequencing indicated that the two main CAFs (extracellular matrix and wound healing CAFs) that produces extracellular matrix could influence the tumor SWE stiffness as well as the antitumor effect of drugs. Further, biomimetic nanoparticles inhibited CAFs function could attenuate tumor hypoxia by increasing proportion of inflammatory blood vessels and oxygen transport capacity. Therefore, our finding is fundamental for understanding the role of CAFs on affecting SWE stiffness and drugs antitumor effects, which can be further implied in the potential clinical theranostic predicting in neoadjuvant therapy efficacy through non-invasive analyzing of SWE imaging.
Keywords: Biomimetic nanoparticles; Cancer-associated fibroblasts; Shear-wave elasticity imaging; Theranostic prediction.
© 2022 The Authors.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures









Similar articles
-
Using Cancer-Associated Fibroblasts as a Shear-Wave Elastography Imaging Biomarker to Predict Anti-PD-1 Efficacy of Triple-Negative Breast Cancer.Int J Mol Sci. 2025 Apr 9;26(8):3525. doi: 10.3390/ijms26083525. Int J Mol Sci. 2025. PMID: 40332007 Free PMC article.
-
Tumor stiffness measured by shear wave elastography correlates with tumor hypoxia as well as histologic biomarkers in breast cancer.Cancer Imaging. 2020 Dec 1;20(1):85. doi: 10.1186/s40644-020-00362-7. Cancer Imaging. 2020. PMID: 33256820 Free PMC article.
-
Elastographic features of triple negative breast cancers.Eur Radiol. 2016 Apr;26(4):1090-7. doi: 10.1007/s00330-015-3925-7. Epub 2015 Aug 1. Eur Radiol. 2016. PMID: 26231093
-
Technical performance of shear wave elastography for measuring liver stiffness in pediatric and adolescent patients: a systematic review and meta-analysis.Eur Radiol. 2019 May;29(5):2560-2572. doi: 10.1007/s00330-018-5900-6. Epub 2019 Jan 7. Eur Radiol. 2019. PMID: 30617493
-
Regulation of heterogeneous cancer-associated fibroblasts: the molecular pathology of activated signaling pathways.J Exp Clin Cancer Res. 2020 Jun 16;39(1):112. doi: 10.1186/s13046-020-01611-0. J Exp Clin Cancer Res. 2020. PMID: 32546182 Free PMC article. Review.
Cited by
-
Correlation between Baseline Conventional Ultrasounds, Shear-Wave Elastography Indicators, and Neoadjuvant Therapy Efficacy in Triple-Negative Breast Cancer.Diagnostics (Basel). 2023 Oct 11;13(20):3178. doi: 10.3390/diagnostics13203178. Diagnostics (Basel). 2023. PMID: 37891999 Free PMC article.
-
Cancer cell membrane-coated nanoparticles: a promising anti-tumor bionic platform.RSC Adv. 2024 Apr 2;14(15):10608-10637. doi: 10.1039/d4ra01026d. eCollection 2024 Mar 26. RSC Adv. 2024. PMID: 38567339 Free PMC article. Review.
-
Using Cancer-Associated Fibroblasts as a Shear-Wave Elastography Imaging Biomarker to Predict Anti-PD-1 Efficacy of Triple-Negative Breast Cancer.Int J Mol Sci. 2025 Apr 9;26(8):3525. doi: 10.3390/ijms26083525. Int J Mol Sci. 2025. PMID: 40332007 Free PMC article.
-
Better together: biomimetic nanomedicines for high performance tumor therapy.Beilstein J Nanotechnol. 2025 Aug 5;16:1246-1276. doi: 10.3762/bjnano.16.92. eCollection 2025. Beilstein J Nanotechnol. 2025. PMID: 40791939 Free PMC article. Review.
-
Advancements in Cell Membrane-Derived Biomimetic Nanotherapeutics for Breast Cancer.Int J Nanomedicine. 2025 May 12;20:6059-6083. doi: 10.2147/IJN.S502144. eCollection 2025. Int J Nanomedicine. 2025. PMID: 40385497 Free PMC article. Review.
References
-
- Sun X., He X., Zhang Y., Hosaka K., Andersson P., Wu J., Wu J., Jing X., Du Q., Hui X., Ding B., Guo Z., Hong A., Liu X., Wang Y., Ji Q., Beyaert R., Yang Y., Li Q., Cao Y.A.-O. Inflammatory cell-derived CXCL3 promotes pancreatic cancer metastasis through a novel myofibroblast-hijacked cancer escape mechanism. Gut. 2022;71(1) - PubMed
-
- Chen X., Song E. Turning foes to friends: targeting cancer-associated fibroblasts, Nature reviews. Drug discovery. 2019;18:99–115. - PubMed
-
- Costa A., Kieffer Y., Scholer-Dahirel A., Pelon F., Bourachot B., Cardon M., Sirven P., Magagna I., Fuhrmann L., Bernard C., Bonneau C., Kondratova M., Kuperstein I., Zinovyev A., Givel A.M., Parrini M.C., Soumelis V., Vincent-Salomon A., Mechta-Grigoriou F. Fibroblast heterogeneity and immunosuppressive environment in human breast cancer. Cancer Cell. 2018;33:463–479. e10. - PubMed
-
- Sun X., Wu B.A.-O.X., Chiang H.A.-O., Deng H., Zhang X., Xiong W., Liu J., Rozeboom A.M., Harris B.T., Blommaert E.A.-O., Gomez A.A.-O.X., Garcia R.A.-O., Zhou Y.A.-O.X., Mitra P., Prevost M., Zhang D., Banik D., Isaacs C., Berry D., Lai C., Chaldekas K., Latham P.A.-O., Brantner C.A., Popratiloff A., Jin V.A.-O., Zhang N.A.-O., Hu Y., Pujana M.A.-O., Curiel T.A.-O., An Z.A.-O., Li R.A.-O. Tumour DDR1 promotes collagen fibre alignment to instigate immune exclusion. Nature. 2021;599:673–678. - PMC - PubMed
-
- Koikawa K., Kibe S., Suizu F., Sekino N., Kim N., Manz T.D., Pinch B.J., Akshinthala D., Verma A., Gaglia G., Nezu Y., Ke S., Qiu C., Ohuchida K., Oda Y., Lee T.H., Wegiel B., Clohessy J.G., London N., Santagata S., Wulf G.M., Hidalgo M., Muthuswamy S.K., Nakamura M., Gray N.S., Zhou X.Z., Lu K.P. Targeting Pin1 renders pancreatic cancer eradicable by synergizing with immunochemotherapy. Cell. 2021;184:4753–4771. - PMC - PubMed
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

