The mutational spectrum of SARS-CoV-2 genomic and antigenomic RNA
- PMID: 36382519
- PMCID: PMC9667359
- DOI: 10.1098/rspb.2022.1747
The mutational spectrum of SARS-CoV-2 genomic and antigenomic RNA
Abstract
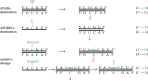
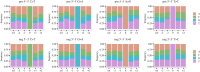
The raw material for viral evolution is provided by intra-host mutations occurring during replication, transcription or post-transcription. Replication and transcription of Coronaviridae proceed through the synthesis of negative-sense 'antigenomes' acting as templates for positive-sense genomic and subgenomic RNA. Hence, mutations in the genomes of SARS-CoV-2 and other coronaviruses can occur during (and after) the synthesis of either negative-sense or positive-sense RNA, with potentially distinct patterns and consequences. We explored for the first time the mutational spectrum of SARS-CoV-2 (sub)genomic and anti(sub)genomic RNA. We use a high-quality deep sequencing dataset produced using a quantitative strand-aware sequencing method, controlled for artefacts and sequencing errors, and scrutinized for accurate detection of within-host diversity. The nucleotide differences between negative- and positive-sense strand consensus vary between patients and do not show dependence on age or sex. Similarities and differences in mutational patterns between within-host minor variants on the two RNA strands suggested strand-specific mutations or editing by host deaminases and oxidative damage. We observe generally neutral and slight negative selection on the negative strand, contrasting with purifying selection in ORF1a, ORF1b and S genes of the positive strand of the genome.
Keywords: RNA editing; SARS-CoV-2; antigenomes; mutational spectrum.
Conflict of interest statement
We declare we have no competing interests.
Figures


References
-
- World Health Organization. 2022. COVID-19 weekly epidemiological update, edition 110, 21 September. https://apps.who.int/iris/bitstream/handle/10665/363125/nCoV-weekly-sitr....
-
- Stern A, Andino R. 2016. Viral evolution: it is all about mutations. In Viral pathogenesis: from basics to systems biology (eds Katze MG, Korth MJ, Law GL, Nathanson N), pp. 233-240. New York, NY: Academic Press.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

