Urinary Microbiome of Reproductive-Age Asymptomatic European Women
- PMID: 36383025
- PMCID: PMC9769847
- DOI: 10.1128/spectrum.01308-22
Urinary Microbiome of Reproductive-Age Asymptomatic European Women
Abstract
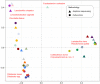
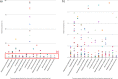
The knowledge of bacterial species diversity within the female urinary microbiome (FUM) is essential for understanding the role of the FUM in urinary tract health and disease. This study aimed to characterize the bacterial species diversity of the FUM of asymptomatic reproductive-age European women by combining extended culturomics and long-read sequencing of the near-full-length 16S rRNA gene. A total of 297 bacterial species (median of 53 species/sample) were identified, yet only 22% of the species were detected by both culture and sequencing methods. Recently recognized Gardnerella, Lactobacillus, and Limosilactobacillus species and 5 new putative Corynebacterium species were identified by culturomics, while anaerobic species (e.g., 11 Peptoniphilus spp.) were mostly detected by amplicon sequencing. Notably, there was not a single species common to all samples, although members of the genus Lactobacillus were detected in all. Lactobacillus crispatus, Lactobacillus iners, and Lactobacillus mulieris were observed in high relative abundance in several samples, as well as other species (e.g., Streptococcus agalactiae, Fannyhessea vaginae, Gardnerella vaginalis, Gardnerella swidsinskii), while low-abundance members (e.g., Finegoldia magna) were often more prevalent. A moderate correlation (Mantel test; r = 0.5) between community structure types captured by culturomics and amplicon sequencing was observed, highlighting the benefit of combining both methodologies. This study provided a detailed FUM structure at the species level, which is critical to unveil the potential relationship between specific microbiome members and urinary diseases/disorders. Moreover, the different capacity to characterize microbiome profiles of culturomic and amplicon sequencing is described, providing valuable insights for further urinary microbiome studies. IMPORTANCE The bacterial species diversity within the female urinary microbiome (FUM) has been insufficiently characterized. This study demonstrated that complementarity between optimized culture-dependent and -independent approaches is highly beneficial for comprehensive FUM species profiling by detecting higher FUM species diversity than previously reported, including identification of unreported species belonging to the genera Lactobacillus, Limosilactobacillus, and Latilactobacillus and putative novel Corynebacterium species. Although some species were present in high relative abundance, low-abundance members were more prevalent. FUM classification into community structure types demonstrated high interindividual differences in urinary microbiome composition among asymptomatic women. We also report moderate correlation between culture-dependent and -independent derived data-highlighting drawbacks of each methodological approach. Our findings suggest that FUM bacterial diversity reported from previous studies may be underestimated. Finally, our results contribute to the fundamental knowledge of the FUM required for further exploration of the urinary microbiome role in urinary tract diseases.
Keywords: 16S rRNA gene amplicon sequencing; Corynebacterium; Gardnerella; Lactobacillus; extended culturomics; midstream urine.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


References
-
- Hilt EE, McKinley K, Pearce MM, Rosenfeld AB, Zilliox MJ, Mueller ER, Brubaker L, Gai X, Wolfe AJ, Schreckenberger PC. 2014. Urine is not sterile: use of enhanced urine culture techniques to detect resident bacterial flora in the adult female bladder. J Clin Microbiol 52:871–876. doi: 10.1128/JCM.02876-13. - DOI - PMC - PubMed
-
- Price TK, Dune T, Hilt EE, Thomas-White KJ, Kliethermes S, Brincat C, Brubaker L, Wolfe AJ, Mueller ER, Schreckenberger PC. 2016. The clinical urine culture: enhanced techniques improve detection of clinically relevant microorganisms. J Clin Microbiol 54:1216–1222. doi: 10.1128/JCM.00044-16. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

