Quantifying the impact of immune history and variant on SARS-CoV-2 viral kinetics and infection rebound: A retrospective cohort study
- PMID: 36383192
- PMCID: PMC9711520
- DOI: 10.7554/eLife.81849
Quantifying the impact of immune history and variant on SARS-CoV-2 viral kinetics and infection rebound: A retrospective cohort study
Abstract
Background: The combined impact of immunity and SARS-CoV-2 variants on viral kinetics during infections has been unclear.
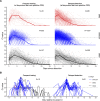
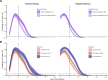
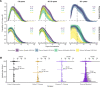
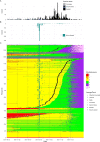
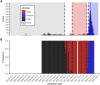
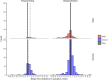
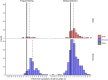
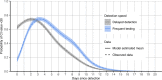
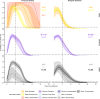
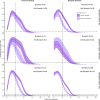
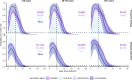
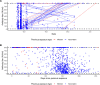
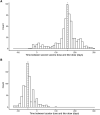
Methods: We characterized 1,280 infections from the National Basketball Association occupational health cohort identified between June 2020 and January 2022 using serial RT-qPCR testing. Logistic regression and semi-mechanistic viral RNA kinetics models were used to quantify the effect of age, variant, symptom status, infection history, vaccination status and antibody titer to the founder SARS-CoV-2 strain on the duration of potential infectiousness and overall viral kinetics. The frequency of viral rebounds was quantified under multiple cycle threshold (Ct) value-based definitions.
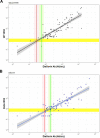
Results: Among individuals detected partway through their infection, 51.0% (95% credible interval [CrI]: 48.3-53.6%) remained potentially infectious (Ct <30) 5 days post detection, with small differences across variants and vaccination status. Only seven viral rebounds (0.7%; N=999) were observed, with rebound defined as 3+days with Ct <30 following an initial clearance of 3+days with Ct ≥30. High antibody titers against the founder SARS-CoV-2 strain predicted lower peak viral loads and shorter durations of infection. Among Omicron BA.1 infections, boosted individuals had lower pre-booster antibody titers and longer clearance times than non-boosted individuals.
Conclusions: SARS-CoV-2 viral kinetics are partly determined by immunity and variant but dominated by individual-level variation. Since booster vaccination protects against infection, longer clearance times for BA.1-infected, boosted individuals may reflect a less effective immune response, more common in older individuals, that increases infection risk and reduces viral RNA clearance rate. The shifting landscape of viral kinetics underscores the need for continued monitoring to optimize isolation policies and to contextualize the health impacts of therapeutics and vaccines.
Funding: Supported in part by CDC contract #200-2016-91779, a sponsored research agreement to Yale University from the National Basketball Association contract #21-003529, and the National Basketball Players Association.
Keywords: COVID-19; SARS-CoV-2; antibody; epidemiology; global health; human; immunity; immunology; inflammation; omicron; viral kinetics; viruses.
© 2022, Hay, Kissler, Fauver et al.
Conflict of interest statement
JH, MN, YH, DH No competing interests declared, SK SMK has a consulting agreement with the NBA, JF, NG has a consulting agreement for Tempus and receives financial support from Tempus to develop SARS-CoV-2 diagnostic tests, CM, CT, RS, SC is an employee of IQVIA, Real World Solutions, DA is co-owner of Infection Control Education for Major Sports, GK, MM, MP, SK, AM, IE, DS, TB, BH, JD, CM is an employee of Tempus Labs, JD is an employee of the NBA, YG has a consulting agreement with the NBA
Figures



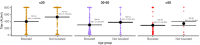

References
-
- Aksamentov I, Neher R. NextClade. The Journal of Open Source Software. 2021;6:3773. doi: 10.21105/joss.03773. - DOI
-
- BaseSpace Labs DRAGEN COVID lineage. 3.5.9Illumina. 2021 https://sapac.illumina.com/products/by-type/informatics-products/basespa...
-
- Boucau J, Marino C, Regan J, Uddin R, Choudhary MC, Flynn JP, Chen G, Stuckwisch AM, Mathews J, Liew MY, Singh A, Lipiner T, Kittilson A, Melberg M, Li Y, Gilbert RF, Reynolds Z, Iyer SL, Chamberlin GC, Vyas TD, Goldberg MB, Vyas JM, Li JZ, Lemieux JE, Siedner MJ, Barczak AK. Duration of shedding of culturable virus in SARS-cov-2 omicron (BA.1) infection. The New England Journal of Medicine. 2022a;387:275–277. doi: 10.1056/NEJMc2202092. - DOI - PMC - PubMed
-
- Boucau J, Uddin R, Marino C, Regan J, Flynn JP, Choudhary MC, Chen G, Stuckwisch AM, Mathews J, Liew MY, Singh A, Reynolds Z, Iyer SL, Chamberlin GC, Vyas TD, Vyas JM, Turbett SE, Li JZ, Lemieux JE, Barczak AK, Siedner MJ. Virologic Characterization of Symptom Rebound Following Nirmatrelvir-Ritonavir Treatment for COVID-19. medRxiv. 2022b doi: 10.1101/2022.05.24.22275326. - DOI - PMC - PubMed
-
- Brihn A, Chang J, OYong K, Balter S, Terashita D, Rubin Z, Yeganeh N. Diagnostic performance of an antigen test with RT-PCR for the detection of SARS-cov-2 in a hospital setting-Los Angeles County, California, june-august 2020. MMWR. Morbidity and Mortality Weekly Report. 2021;70:702–706. doi: 10.15585/mmwr.mm7019a3. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Supplementary concepts
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

