Cryo-EM reveals the conformational epitope of human monoclonal antibody PAM1.4 broadly reacting with polymorphic malarial protein VAR2CSA
- PMID: 36383559
- PMCID: PMC9668162
- DOI: 10.1371/journal.ppat.1010924
Cryo-EM reveals the conformational epitope of human monoclonal antibody PAM1.4 broadly reacting with polymorphic malarial protein VAR2CSA
Abstract
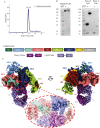
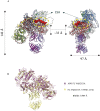
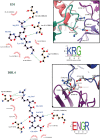
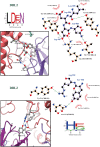
Malaria during pregnancy is a major global health problem caused by infection with Plasmodium falciparum parasites. Severe effects arise from the accumulation of infected erythrocytes in the placenta. Here, erythrocytes infected by late blood-stage parasites adhere to placental chondroitin sulphate A (CS) via VAR2CSA-type P. falciparum erythrocyte membrane protein 1 (PfEMP1) adhesion proteins. Immunity to placental malaria is acquired through exposure and mediated through antibodies to VAR2CSA. Through evolution, the VAR2CSA proteins have diversified in sequence to escape immune recognition but retained their overall macromolecular structure to maintain CS binding affinity. This structural conservation may also have allowed development of broadly reactive antibodies to VAR2CSA in immune women. Here we show the negative stain and cryo-EM structure of the only known broadly reactive human monoclonal antibody, PAM1.4, in complex with VAR2CSA. The data shows how PAM1.4's broad VAR2CSA reactivity is achieved through interactions with multiple conserved residues of different sub-domains forming conformational epitope distant from the CS binding site on the VAR2CSA core structure. Thus, while PAM1.4 may represent a class of antibodies mediating placental malaria immunity by inducing phagocytosis or NK cell-mediated cytotoxicity, it is likely that broadly CS binding-inhibitory antibodies target other epitopes at the CS binding site. Insights on both types of broadly reactive monoclonal antibodies may aid the development of a vaccine against placental malaria.
Copyright: © 2022 Raghavan et al. This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Conflict of interest statement
We have read the journal’s policy and the authors of this manuscript have the following competing interests: AS, and MAN are listed as co-inventors on a patent family covering the use of VAR2CSA to target and diagnose cancer. AS and is listed as co-inventor on a patent on using VAR2CSA as a prophylactic malaria vaccine during pregnancy. The other authors have no conflicts of interest.
Figures

References
-
- World Malaria Report 2021. 2021.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials

