Proteogenomic analysis of lung adenocarcinoma reveals tumor heterogeneity, survival determinants, and therapeutically relevant pathways
- PMID: 36384096
- PMCID: PMC9729884
- DOI: 10.1016/j.xcrm.2022.100819
Proteogenomic analysis of lung adenocarcinoma reveals tumor heterogeneity, survival determinants, and therapeutically relevant pathways
Abstract
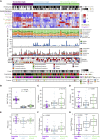
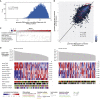
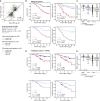
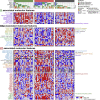
We present a deep proteogenomic profiling study of 87 lung adenocarcinoma (LUAD) tumors from the United States, integrating whole-genome sequencing, transcriptome sequencing, proteomics and phosphoproteomics by mass spectrometry, and reverse-phase protein arrays. We identify three subtypes from somatic genome signature analysis, including a transition-high subtype enriched with never smokers, a transversion-high subtype enriched with current smokers, and a structurally altered subtype enriched with former smokers, TP53 alterations, and genome-wide structural alterations. We show that within-tumor correlations of RNA and protein expression associate with tumor purity and immune cell profiles. We detect and independently validate expression signatures of RNA and protein that predict patient survival. Additionally, among co-measured genes, we found that protein expression is more often associated with patient survival than RNA. Finally, integrative analysis characterizes three expression subtypes with divergent mutations, proteomic regulatory networks, and therapeutic vulnerabilities. This proteogenomic characterization provides a foundation for molecularly informed medicine in LUAD.
Keywords: TP53; cancer; immune; lung adenocarcinoma; proteogenomics; proteomics; subtype; survival; transcriptomics; whole genome.
Published by Elsevier Inc.
Conflict of interest statement
Declaration of interests M.D.W., R.F.B., and C.D.S. are inventors for a provisional patent application related to findings reported in this manuscript. J.S.H.L. serves as Chief Science and Innovation Officer for Ellison Institute, LLC (paid); board of trustee for Health and Environmental Institute, Inc. (unpaid, travel support); and scientific advisory board for AtlasXomics, Inc., and ATOM, Inc. (unpaid, travel support).
Figures



References
-
- Lin J., Kamamia C., Brown D., Shao S., McGlynn K.A., Nations J.A., Carter C.A., Shriver C.D., Zhu K. Survival among lung cancer patients in the U.S. Military Health system: a comparison with the SEER population. Cancer Epidemiol. Biomarkers Prev. 2018;27:673–679. doi: 10.1158/1055-9965.EPI-17-0822. - DOI - PMC - PubMed
-
- Grilley-Olson J.E., Hayes D.N., Moore D.T., Leslie K.O., Wilkerson M.D., Qaqish B.F., Hayward M.C., Cabanski C.R., Yin X., Socinski M.A., et al. Validation of interobserver agreement in lung cancer assessment: hematoxylin-eosin diagnostic reproducibility for non-small cell lung cancer: the 2004 World Health Organization classification and therapeutically relevant subsets. Arch. Pathol. Lab Med. 2013;137:32–40. doi: 10.5858/arpa.2012-0033-OA. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

