IKKβ primes inflammasome formation by recruiting NLRP3 to the trans-Golgi network
- PMID: 36384135
- PMCID: PMC7614333
- DOI: 10.1016/j.immuni.2022.10.021
IKKβ primes inflammasome formation by recruiting NLRP3 to the trans-Golgi network
Abstract
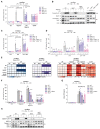
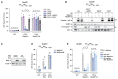
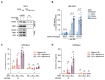
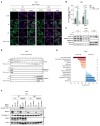
The NLRP3 inflammasome plays a central role in antimicrobial defense as well as in the context of sterile inflammatory conditions. NLRP3 activity is governed by two independent signals: the first signal primes NLRP3, rendering it responsive to the second signal, which then triggers inflammasome formation. Our understanding of how NLRP3 priming contributes to inflammasome activation remains limited. Here, we show that IKKβ, a kinase activated during priming, induces recruitment of NLRP3 to phosphatidylinositol-4-phosphate (PI4P), a phospholipid enriched on the trans-Golgi network. NEK7, a mitotic spindle kinase that had previously been thought to be indispensable for NLRP3 activation, was redundant for inflammasome formation when IKKβ recruited NLRP3 to PI4P. Studying iPSC-derived human macrophages revealed that the IKKβ-mediated NEK7-independent pathway constitutes the predominant NLRP3 priming mechanism in human myeloid cells. Our results suggest that PI4P binding represents a primed state into which NLRP3 is brought by IKKβ activity.
Keywords: NEK7; NLRP3; iPSCs; inflammasome; innate immunity; macrophages; monocytes; pattern recognition.
Copyright © 2022 Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests The authors declare no competing interests.
Figures


Comment in
-
Humans pIKK-up NLRP3 to skip NEK7.Trends Immunol. 2022 Dec;43(12):947-949. doi: 10.1016/j.it.2022.10.007. Epub 2022 Nov 17. Trends Immunol. 2022. PMID: 36404209
-
IKKβ ignites the inflammasome.Nat Rev Immunol. 2023 Jan;23(1):6. doi: 10.1038/s41577-022-00818-w. Nat Rev Immunol. 2023. PMID: 36470972 No abstract available.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

