GPER1 links estrogens to centrosome amplification and chromosomal instability in human colon cells
- PMID: 36384894
- PMCID: PMC9670797
- DOI: 10.26508/lsa.202201499
GPER1 links estrogens to centrosome amplification and chromosomal instability in human colon cells
Abstract
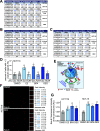
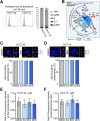
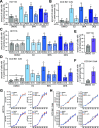
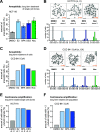
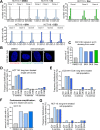
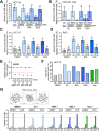
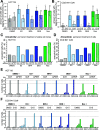
The role of the alternate G protein-coupled estrogen receptor 1 (GPER1) in colorectal cancer (CRC) development and progression is unclear, not least because of conflicting clinical and experimental evidence for pro- and anti-tumorigenic activities. Here, we show that low concentrations of the estrogenic GPER1 ligands, 17β-estradiol, bisphenol A, and diethylstilbestrol cause the generation of lagging chromosomes in normal colon and CRC cell lines, which manifest in whole chromosomal instability and aneuploidy. Mechanistically, (xeno)estrogens triggered centrosome amplification by inducing centriole overduplication that leads to transient multipolar mitotic spindles, chromosome alignment defects, and mitotic laggards. Remarkably, we could demonstrate a significant role of estrogen-activated GPER1 in centrosome amplification and increased karyotype variability. Indeed, both gene-specific knockdown and inhibition of GPER1 effectively restored normal centrosome numbers and karyotype stability in cells exposed to 17β-estradiol, bisphenol A, or diethylstilbestrol. Thus, our results reveal a novel link between estrogen-activated GPER1 and the induction of key CRC-prone lesions, supporting a pivotal role of the alternate estrogen receptor in colon neoplastic transformation and tumor progression.
© 2022 Bühler et al.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures





References
-
- Kayser G, Gerlach U, Walch A, Nitschke R, Haxelmans S, Kayser K, Hopt U, Werner M, Lassmann S (2005) Numerical and structural centrosome aberrations are an early and stable event in the adenoma-carcinoma sequence of colorectal carcinomas. Virchows Arch 447: 61–65. 10.1007/s00428-004-1191-1 - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
