Compensatory epistasis maintains ACE2 affinity in SARS-CoV-2 Omicron BA.1
- PMID: 36384919
- PMCID: PMC9668218
- DOI: 10.1038/s41467-022-34506-z
Compensatory epistasis maintains ACE2 affinity in SARS-CoV-2 Omicron BA.1
Abstract
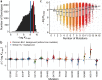
The Omicron BA.1 variant emerged in late 2021 and quickly spread across the world. Compared to the earlier SARS-CoV-2 variants, BA.1 has many mutations, some of which are known to enable antibody escape. Many of these antibody-escape mutations individually decrease the spike receptor-binding domain (RBD) affinity for ACE2, but BA.1 still binds ACE2 with high affinity. The fitness and evolution of the BA.1 lineage is therefore driven by the combined effects of numerous mutations. Here, we systematically map the epistatic interactions between the 15 mutations in the RBD of BA.1 relative to the Wuhan Hu-1 strain. Specifically, we measure the ACE2 affinity of all possible combinations of these 15 mutations (215 = 32,768 genotypes), spanning all possible evolutionary intermediates from the ancestral Wuhan Hu-1 strain to BA.1. We find that immune escape mutations in BA.1 individually reduce ACE2 affinity but are compensated by epistatic interactions with other affinity-enhancing mutations, including Q498R and N501Y. Thus, the ability of BA.1 to evade immunity while maintaining ACE2 affinity is contingent on acquiring multiple interacting mutations. Our results implicate compensatory epistasis as a key factor driving substantial evolutionary change for SARS-CoV-2 and are consistent with Omicron BA.1 arising from a chronic infection.
© 2022. The Author(s).
Conflict of interest statement
A.M.P. and M.M.D. have or have recently consulted for Leyden Labs. J.D.B. has or has recently consulted for Apriori Bio, Oncorus, Moderna, and Merck. J.D.B., A.J.G., and T.N.S. are inventors on Fred Hutch licensed patents related to viral deep mutational scanning. The other authors declare no competing interests.
Figures

References
-
- Liu L, et al. Striking antibody evasion manifested by the Omicron variant of SARS-CoV-2. Nature. 2022;602:676–681. - PubMed
Publication types
MeSH terms
Substances
Supplementary concepts
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

