Global disparities in SARS-CoV-2 genomic surveillance
- PMID: 36385137
- PMCID: PMC9667854
- DOI: 10.1038/s41467-022-33713-y
Global disparities in SARS-CoV-2 genomic surveillance
Abstract
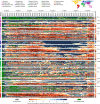
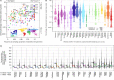
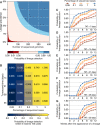
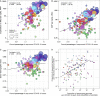
Genomic sequencing is essential to track the evolution and spread of SARS-CoV-2, optimize molecular tests, treatments, vaccines, and guide public health responses. To investigate the global SARS-CoV-2 genomic surveillance, we used sequences shared via GISAID to estimate the impact of sequencing intensity and turnaround times on variant detection in 189 countries. In the first two years of the pandemic, 78% of high-income countries sequenced >0.5% of their COVID-19 cases, while 42% of low- and middle-income countries reached that mark. Around 25% of the genomes from high income countries were submitted within 21 days, a pattern observed in 5% of the genomes from low- and middle-income countries. We found that sequencing around 0.5% of the cases, with a turnaround time <21 days, could provide a benchmark for SARS-CoV-2 genomic surveillance. Socioeconomic inequalities undermine the global pandemic preparedness, and efforts must be made to support low- and middle-income countries improve their local sequencing capacity.
© 2022. The Author(s).
Conflict of interest statement
N.D.G. is an infectious diseases consultant for Tempus Labs and the National Basketball Association. M.A.S. receives grants and contracts from the National Institutes of Health, the US Food & Drug Administration, the US Department of Veterans Affairs and Janssen Research & Development. O.G.P. has undertaken work for AstraZeneca on SARS-CoV-2 classification and genetic lineage nomenclature. The remaining authors declare no competing interests.
Figures
Update of
-
Global disparities in SARS-CoV-2 genomic surveillance.medRxiv [Preprint]. 2021 Dec 9:2021.08.21.21262393. doi: 10.1101/2021.08.21.21262393. medRxiv. 2021. Update in: Nat Commun. 2022 Nov 16;13(1):7003. doi: 10.1038/s41467-022-33713-y. PMID: 34462754 Free PMC article. Updated. Preprint.
References
-
- WHO. WHO COVID-19 Explorer. https://worldhealthorg.shinyapps.io/covid/ (2021).
-
- Rydland HT, Friedman J, Stringhini S, Link BG, Eikemo TA. The radically unequal distribution of Covid-19 vaccinations: a predictable yet avoidable symptom of the fundamental causes of inequality. Humanit. Soc. Sci. Commun. 2022;9:1–6. doi: 10.1057/s41599-022-01073-z. - DOI
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

