Functional and clinical studies reveal pathophysiological complexity of CLCN4-related neurodevelopmental condition
- PMID: 36385166
- PMCID: PMC9908558
- DOI: 10.1038/s41380-022-01852-9
Functional and clinical studies reveal pathophysiological complexity of CLCN4-related neurodevelopmental condition
Abstract
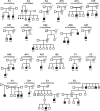
Missense and truncating variants in the X-chromosome-linked CLCN4 gene, resulting in reduced or complete loss-of-function (LOF) of the encoded chloride/proton exchanger ClC-4, were recently demonstrated to cause a neurocognitive phenotype in both males and females. Through international clinical matchmaking and interrogation of public variant databases we assembled a database of 90 rare CLCN4 missense variants in 90 families: 41 unique and 18 recurrent variants in 49 families. For 43 families, including 22 males and 33 females, we collated detailed clinical and segregation data. To confirm causality of variants and to obtain insight into disease mechanisms, we investigated the effect on electrophysiological properties of 59 of the variants in Xenopus oocytes using extended voltage and pH ranges. Detailed analyses revealed new pathophysiological mechanisms: 25% (15/59) of variants demonstrated LOF, characterized by a "shift" of the voltage-dependent activation to more positive voltages, and nine variants resulted in a toxic gain-of-function, associated with a disrupted gate allowing inward transport at negative voltages. Functional results were not always in line with in silico pathogenicity scores, highlighting the complexity of pathogenicity assessment for accurate genetic counselling. The complex neurocognitive and psychiatric manifestations of this condition, and hitherto under-recognized impacts on growth, gastrointestinal function, and motor control are discussed. Including published cases, we summarize features in 122 individuals from 67 families with CLCN4-related neurodevelopmental condition and suggest future research directions with the aim of improving the integrated care for individuals with this diagnosis.
© 2022. The Author(s).
Conflict of interest statement
SW received consultancy fees from UCB, Biocodex, Xenon Pharmaceuticals, Zogenix, Lundbeck, Knopp Biosciences, and Encoded Therapeutics. KMc, TB, and ET are employees of GeneDx. ZP is an employee of Ambry. CB is an employee of Centogene, GmbH. The Department of Molecular and Human Genetics at Baylor College of Medicine receives revenue from clinical genetic testing completed at Baylor Genetics Laboratories. All other authors declare no competing interests.
Figures



References
-
- Raynaud M, Gendrot C, Dessay B, Moncla A, Ayrault AD, Moizard MP, et al. X-linked mental retardation with neonatal hypotonia in a French family (MRX15): gene assignment to Xp11.22-Xp21.1. Am J Med Genet. 1996;64:97–106. doi: 10.1002/(SICI)1096-8628(19960712)64:1<97::AID-AJMG17>3.0.CO;2-N. - DOI - PubMed
-
- Claes S, Vogels A, Holvoet M, Devriendt K, Raeymaekers P, Cassiman JJ, et al. Regional localization of two genes for nonspecific X-linked mental retardation to Xp22.3-p22.2 (MRX49) and Xp11.3-p11.21 (MRX50) Am J Med Genet. 1997;73:474–9. doi: 10.1002/(SICI)1096-8628(19971231)73:4<474::AID-AJMG18>3.0.CO;2-O. - DOI - PubMed
-
- Palmer EE, Stuhlmann T, Weinert S, Haan E, Van Esch H, Holvoet M, et al. De novo and inherited mutations in the X-linked gene CLCN4 are associated with syndromic intellectual disability and behavior and seizure disorders in males and females. Mol Psychiatry. 2018;23:222–30. doi: 10.1038/mp.2016.135. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

