Direct capture and amplification of nucleic acids using a universal, elution-free magnetic bead-based method for rapid pathogen detection in multiple types of biological samples
- PMID: 36385304
- PMCID: PMC9668711
- DOI: 10.1007/s00216-022-04422-8
Direct capture and amplification of nucleic acids using a universal, elution-free magnetic bead-based method for rapid pathogen detection in multiple types of biological samples
Abstract
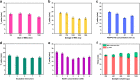
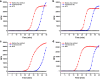
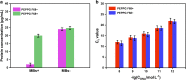
Nucleic acid amplification tests (NAATs) have become an attractive approach for pathogen detection, and obtaining high-quality nucleic acid extracts from biological samples plays a critical role in ensuring accurate NAATs. In this work, we established an elution-free magnetic bead (MB)-based method by introducing polyethylene-polypropylene glycol (PEPPG) F68 in lysis buffer and using NaOH solution instead of alcohols as the washing buffer for rapid nucleic acid extraction from multiple types of biological samples, including nasopharyngeal swabs, serum, milk, and pork, which bypassed the nucleic acid elution step and allowed the nucleic acid/MB composite to be directly used as the template for amplification reactions. The entire extraction process was able to be completed in approximately 7 min. Even though the nucleic acid/MB composite could not be used for quantitative real-time PCR (qPCR) assays, this elution-free MB-based method significantly improved the sensitivity of the loop-mediated isothermal amplification (LAMP) assay. The sensitivity of the quantitative real-time LAMP (qLAMP) assays combined with this elution-free MB-based method showed an improvement of one to three orders of magnitude compared with qLAMP or qPCR assays combined with the traditional MB-based method. In addition to manual operation, like the traditional MB-based method, this universal, rapid, and facile nucleic acid extraction method also has potential for integration into automated robotic processing, making it particularly suitable for the establishment of an analysis platform for ultrafast and sensitive pathogen detection in various biological samples both in centralized laboratories and at remote sites.
Keywords: Elution-free; Magnetic beads; Nucleic acid amplification tests; Nucleic acid extraction; Pathogen detection; Polyethylene-polypropylene glycol F68.
© 2022. Springer-Verlag GmbH Germany, part of Springer Nature.
Conflict of interest statement
There are no conflicts of interest to declare.
Figures



Similar articles
-
Limited-resource preparable chitosan magnetic particles for extracting amplification-ready nucleic acid from complex biofluids.Analyst. 2021 Dec 20;147(1):165-177. doi: 10.1039/d1an01150b. Analyst. 2021. PMID: 34870658
-
Direct loop mediated isothermal amplification on filters for quantification of Dehalobacter in groundwater.J Microbiol Methods. 2016 Dec;131:61-67. doi: 10.1016/j.mimet.2016.09.025. Epub 2016 Oct 5. J Microbiol Methods. 2016. PMID: 27720723 Free PMC article.
-
Highly sensitive detection of gluten-containing cereals in food samples by real-time Loop-mediated isothermal AMPlification (qLAMP) and real-time polymerase chain reaction (qPCR).Food Chem. 2018 Apr 25;246:156-163. doi: 10.1016/j.foodchem.2017.11.005. Epub 2017 Nov 6. Food Chem. 2018. PMID: 29291834
-
Chemical Trends in Sample Preparation for Nucleic Acid Amplification Testing (NAAT): A Review.Biosensors (Basel). 2023 Nov 10;13(11):980. doi: 10.3390/bios13110980. Biosensors (Basel). 2023. PMID: 37998155 Free PMC article. Review.
-
Emerging ultrafast nucleic acid amplification technologies for next-generation molecular diagnostics.Biosens Bioelectron. 2019 Sep 15;141:111448. doi: 10.1016/j.bios.2019.111448. Epub 2019 Jun 18. Biosens Bioelectron. 2019. PMID: 31252258 Review.
Cited by
-
Research Progress of Nucleic Acid Detection Technology for Genetically Modified Maize.Int J Mol Sci. 2023 Jul 31;24(15):12247. doi: 10.3390/ijms241512247. Int J Mol Sci. 2023. PMID: 37569623 Free PMC article. Review.
-
The Rapid Detection of Salmonella enterica, Listeria monocytogenes, and Staphylococcus aureus via Polymerase Chain Reaction Combined with Magnetic Beads and Capillary Electrophoresis.Foods. 2023 Oct 24;12(21):3895. doi: 10.3390/foods12213895. Foods. 2023. PMID: 37959014 Free PMC article.
References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

