Colon tumour cell death causes mTOR dependence by paracrine P2X4 stimulation
- PMID: 36385525
- PMCID: PMC7613947
- DOI: 10.1038/s41586-022-05426-1
Colon tumour cell death causes mTOR dependence by paracrine P2X4 stimulation
Abstract
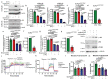
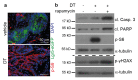
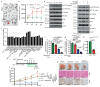
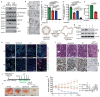
Solid cancers exhibit a dynamic balance between cell death and proliferation ensuring continuous tumour maintenance and growth1,2. Increasing evidence links enhanced cancer cell apoptosis to paracrine activation of cells in the tumour microenvironment initiating tissue repair programs that support tumour growth3,4, yet the direct effects of dying cancer cells on neighbouring tumour epithelia and how this paracrine effect potentially contributes to therapy resistance are unclear. Here we demonstrate that chemotherapy-induced tumour cell death in patient-derived colorectal tumour organoids causes ATP release triggering P2X4 (also known as P2RX4) to mediate an mTOR-dependent pro-survival program in neighbouring cancer cells, which renders surviving tumour epithelia sensitive to mTOR inhibition. The induced mTOR addiction in persisting epithelial cells is due to elevated production of reactive oxygen species and subsequent increased DNA damage in response to the death of neighbouring cells. Accordingly, inhibition of the P2X4 receptor or direct mTOR blockade prevents induction of S6 phosphorylation and synergizes with chemotherapy to cause massive cell death induced by reactive oxygen species and marked tumour regression that is not seen when individually applied. Conversely, scavenging of reactive oxygen species prevents cancer cells from becoming reliant on mTOR activation. Collectively, our findings show that dying cancer cells establish a new dependency on anti-apoptotic programs in their surviving neighbours, thereby creating an opportunity for combination therapy in P2X4-expressing epithelial tumours.
© 2022. The Author(s), under exclusive licence to Springer Nature Limited.
Conflict of interest statement
M.S., J.G. and F.R.G. filed a patent regarding the use of P2X4 inhibitors in combination with cytotoxic compounds. F.J.d.S is employee of Genentech and own Roche shares. F.R.G. is a consultant for Amazentis not related to this study. All other authors declare no competing financial interests.
Figures


Comment in
-
ATP released from dying cancer cells stimulates P2X4 receptors and mTOR in their neighbors.Purinergic Signal. 2024 Feb;20(1):1-4. doi: 10.1007/s11302-023-09926-1. Epub 2023 Feb 8. Purinergic Signal. 2024. PMID: 36750529 Free PMC article. No abstract available.
References
-
- Diwanji N, Bergmann A. Two Sides of the Same Coin-Compensatory Proliferation in Regeneration and Cancer. Adv Exp Med Biol. 2019;1167:65–85. - PubMed
-
- Fonseca BD, Smith EM, Lee VH, MacKintosh C, Proud CG. PRAS40 is a target for mammalian target of rapamycin complex 1 and is required for signaling downstream of this complex. J Biol Chem. 2007;282:24514–24524. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

