Enterococci enhance Clostridioides difficile pathogenesis
- PMID: 36385534
- PMCID: PMC9691601
- DOI: 10.1038/s41586-022-05438-x
Enterococci enhance Clostridioides difficile pathogenesis
Abstract
Enteric pathogens are exposed to a dynamic polymicrobial environment in the gastrointestinal tract1. This microbial community has been shown to be important during infection, but there are few examples illustrating how microbial interactions can influence the virulence of invading pathogens2. Here we show that expansion of a group of antibiotic-resistant, opportunistic pathogens in the gut-the enterococci-enhances the fitness and pathogenesis of Clostridioides difficile. Through a parallel process of nutrient restriction and cross-feeding, enterococci shape the metabolic environment in the gut and reprogramme C. difficile metabolism. Enterococci provide fermentable amino acids, including leucine and ornithine, which increase C. difficile fitness in the antibiotic-perturbed gut. Parallel depletion of arginine by enterococci through arginine catabolism provides a metabolic cue for C. difficile that facilitates increased virulence. We find evidence of microbial interaction between these two pathogenic organisms in multiple mouse models of infection and patients infected with C. difficile. These findings provide mechanistic insights into the role of pathogenic microbiota in the susceptibility to and the severity of C. difficile infection.
© 2022. The Author(s), under exclusive licence to Springer Nature Limited.
Conflict of interest statement
Figures



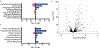




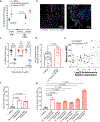
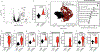

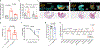
Comment in
-
Helping C. difficile to thrive.Nat Rev Microbiol. 2023 Feb;21(2):65. doi: 10.1038/s41579-022-00838-2. Nat Rev Microbiol. 2023. PMID: 36460929 No abstract available.
References
Methods References
Publication types
MeSH terms
Substances
Grants and funding
- UL1 TR000445/TR/NCATS NIH HHS/United States
- R01 AI145992/AI/NIAID NIH HHS/United States
- R01 AI138581/AI/NIAID NIH HHS/United States
- R35 GM138369/GM/NIGMS NIH HHS/United States
- R01 AI165519/AI/NIAID NIH HHS/United States
- T32 GM071339/GM/NIGMS NIH HHS/United States
- K23 AI121485/AI/NIAID NIH HHS/United States
- R01 HD090061/HD/NICHD NIH HHS/United States
- T32 GM007229/GM/NIGMS NIH HHS/United States
- R01 AI127472/AI/NIAID NIH HHS/United States
- R01 AI164587/AI/NIAID NIH HHS/United States
- P30 AI045008/AI/NIAID NIH HHS/United States
- R01 AT010253/AT/NCCIH NIH HHS/United States
- DP1 DA051620/DA/NIDA NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

