The dynamic lung microbiome in health and disease
- PMID: 36385637
- PMCID: PMC9668228
- DOI: 10.1038/s41579-022-00821-x
The dynamic lung microbiome in health and disease
Abstract
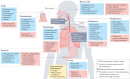
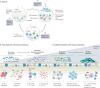
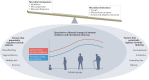
New methods and technologies within the field of lung biology are beginning to shed new light into the microbial world of the respiratory tract. Long considered to be a sterile environment, it is now clear that the human lungs are frequently exposed to live microbes and their by-products. The nature of the lung microbiome is quite distinct from other microbial communities inhabiting our bodies such as those in the gut. Notably, the microbiome of the lung exhibits a low biomass and is dominated by dynamic fluxes of microbial immigration and clearance, resulting in a bacterial burden and microbiome composition that is fluid in nature rather than fixed. As our understanding of the microbial ecology of the lung improves, it is becoming increasingly apparent that certain disease states can disrupt the microbial-host interface and ultimately affect disease pathogenesis. In this Review, we provide an overview of lower airway microbial dynamics in health and disease and discuss future work that is required to uncover novel therapeutic targets to improve lung health.
© 2022. Springer Nature Limited.
Conflict of interest statement
All authors declare no competing interests.
Figures


References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials

