Identification and characterization of alternative sites and molecular probes for SARS-CoV-2 target proteins
- PMID: 36385993
- PMCID: PMC9659918
- DOI: 10.3389/fchem.2022.1017394
Identification and characterization of alternative sites and molecular probes for SARS-CoV-2 target proteins
Abstract
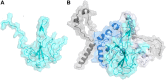
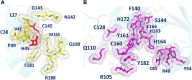
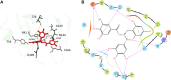
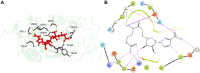
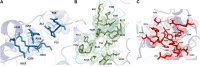
Three protein targets from SARS-CoV-2, the viral pathogen that causes COVID-19, are studied: the main protease, the 2'-O-RNA methyltransferase, and the nucleocapsid (N) protein. For the main protease, the nucleophilicity of the catalytic cysteine C145 is enabled by coupling to three histidine residues, H163 and H164 and catalytic dyad partner H41. These electrostatic couplings enable significant population of the deprotonated state of C145. For the RNA methyltransferase, the catalytic lysine K6968 that serves as a Brønsted base has significant population of its deprotonated state via strong coupling with K6844 and Y6845. For the main protease, Partial Order Optimum Likelihood (POOL) predicts two clusters of biochemically active residues; one includes the catalytic H41 and C145 and neighboring residues. The other surrounds a second pocket adjacent to the catalytic site and includes S1 residues F140, L141, H163, E166, and H172 and also S2 residue D187. This secondary recognition site could serve as an alternative target for the design of molecular probes. From in silico screening of library compounds, ligands with predicted affinity for the secondary site are reported. For the NSP16-NSP10 complex that comprises the RNA methyltransferase, three different sites are predicted. One is the catalytic core at the conserved K-D-K-E motif that includes catalytic residues D6928, K6968, and E7001 plus K6844. The second site surrounds the catalytic core and consists of Y6845, C6849, I6866, H6867, F6868, V6894, D6895, D6897, I6926, S6927, Y6930, and K6935. The third is located at the heterodimer interface. Ligands predicted to have high affinity for the first or second sites are reported. Three sites are also predicted for the nucleocapsid protein. This work uncovers key interactions that contribute to the function of the three viral proteins and also suggests alternative sites for ligand design.
Keywords: Nucleocapsid; POOL; RNA methyltransferase; SARS-CoV-2; coupled amino acids; main protease; secondary sites.
Copyright © 2022 Iyengar, Barnsley, Vu, Bongalonta, Herrod, Scott and Ondrechen.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures

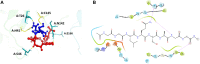

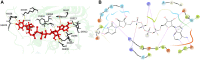
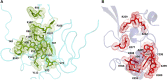
References
LinkOut - more resources
Full Text Sources
Miscellaneous

