Immunotherapy response and microenvironment provide biomarkers of immunotherapy options for patients with lung adenocarcinoma
- PMID: 36386793
- PMCID: PMC9640754
- DOI: 10.3389/fgene.2022.1047435
Immunotherapy response and microenvironment provide biomarkers of immunotherapy options for patients with lung adenocarcinoma
Abstract
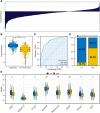
Background: Immunotherapy has been a promising approach option for lung cancer. Method: All the open-accessed data was obtained from the Cancer Genome Atlas (TCGA) database. All the analysis was conducted using the R software analysis. Results: Firstly, the genes differentially expressed in lung cancer immunotherapy responders and non-responders were identified. Then, the lung adenocarcinoma immunotherapy-related genes were determined by LASSO logistic regression and SVM-RFE, respectively. A total of 18 immunotherapy response-related genes were included in our investigation. Subsequently, we constructed the logistics score model. Patients with high logistics score had a better clinical effect on immunotherapy, with 63.2% of patients responding to immunotherapy, while only 12.1% of patients in the low logistics score group responded to immunotherapy. Moreover, we found that pathways related to immunotherapy were mainly enriched in metabolic pathways such as fatty acid metabolism, bile acid metabolism, oxidative phosphorylation, and carcinogenic pathways such as KRAS signaling. Logistics score was positively correlated with NK cells activated, Mast cells resting, Monocytes, Macrophages M2, dendritic cells resting, dendritic cells activated and eosinophils, while was negatively related to Tregs, macrophages M0, macrophages M1, and mast cells activated. In addition, ERVH48-1 was screened for single-cell exploration. The expression of ERVH48-1 increased in patients with distant metastasis, and ERVH48-1 was associated with pathways such as pancreas beta cells, spermatogenesis, G2M checkpoints and KRAS signaling. The result of quantitative real-time PCR showed that ERVH48-1 was upregulated in lung cancer cells. Conclusion: Our study developed an effective signature to predict the immunotherapy response of lung cancer patients.
Keywords: ERVH48-1; immunotherapy response; lung adenocarcinoma; predictive model; tumor environment.
Copyright © 2022 Zhan, Feng, Zhou, Liao, Zhao, Yang, Tan and Shen.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures






References
LinkOut - more resources
Full Text Sources
Miscellaneous

