Natural variation and domestication selection of ZmSULTR3;4 is associated with maize lateral root length in response to salt stress
- PMID: 36388478
- PMCID: PMC9644038
- DOI: 10.3389/fpls.2022.992799
Natural variation and domestication selection of ZmSULTR3;4 is associated with maize lateral root length in response to salt stress
Abstract
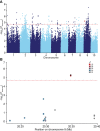
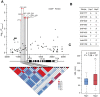
Soil salinity is a major constraint that restricts crop productivity worldwide. Lateral roots (LRs) are important for water and nutrient acquisition, therefore understanding the genetic basis of natural variation in lateral root length (LRL) is of great agronomic relevance to improve salt tolerance in cultivated germplasms. Here, using a genome-wide association study, we showed that the genetic variation in ZmSULTR3;4, which encodes a plasma membrane-localized sulfate transporter, is associated with natural variation in maize LRL under salt stress. The transcript of ZmSULTR3;4 was found preferentially in the epidermal and vascular tissues of root and increased by salt stress, supporting its essential role in the LR formation under salt stress. Further candidate gene association analysis showed that DNA polymorphisms in the promoter region differentiate the expression of ZmSULTR3;4 among maize inbred lines that may contribute to the natural variation of LRL under salt stress. Nucleotide diversity and neutrality tests revealed that ZmSULTR3;4 has undergone selection during maize domestication and improvement. Overall, our results revealed a regulatory role of ZmSULTR3;4 in salt regulated LR growth and uncovered favorable alleles of ZmSULTR3;4, providing an important selection target for breeding salt-tolerant maize cultivar.
Keywords: ZmSULTR3;4; domestication selection; lateral root length; maize (Zea mays); natural variation; salt stress.
Copyright © 2022 Zhang, Zhu, Li, Jia, Wang, Liu, Yang, Chen, Wang, Guo and Li.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures




Similar articles
-
Natural variation in ZmNAC087 contributes to total root length regulation in maize seedlings under salt stress.BMC Plant Biol. 2023 Aug 14;23(1):392. doi: 10.1186/s12870-023-04393-7. BMC Plant Biol. 2023. PMID: 37580686 Free PMC article.
-
The SULTR gene family in maize (Zea mays L.): Gene cloning and expression analyses under sulfate starvation and abiotic stress.J Plant Physiol. 2018 Jan;220:24-33. doi: 10.1016/j.jplph.2017.10.010. Epub 2017 Nov 10. J Plant Physiol. 2018. PMID: 29145069
-
A retrotransposon in an HKT1 family sodium transporter causes variation of leaf Na+ exclusion and salt tolerance in maize.New Phytol. 2018 Feb;217(3):1161-1176. doi: 10.1111/nph.14882. Epub 2017 Nov 15. New Phytol. 2018. PMID: 29139111
-
Genomic screening for artificial selection during domestication and improvement in maize.Ann Bot. 2007 Nov;100(5):967-73. doi: 10.1093/aob/mcm173. Epub 2007 Aug 18. Ann Bot. 2007. PMID: 17704539 Free PMC article. Review.
-
Using Ancient Traits to Convert Soil Health into Crop Yield: Impact of Selection on Maize Root and Rhizosphere Function.Front Plant Sci. 2016 Mar 30;7:373. doi: 10.3389/fpls.2016.00373. eCollection 2016. Front Plant Sci. 2016. PMID: 27066028 Free PMC article. Review.
Cited by
-
Dynamic molecular regulation of salt stress responses in maize (Zea mays L.) seedlings.Front Plant Sci. 2025 Feb 25;16:1535943. doi: 10.3389/fpls.2025.1535943. eCollection 2025. Front Plant Sci. 2025. PMID: 40070712 Free PMC article.
-
Natural variation in ZmNAC087 contributes to total root length regulation in maize seedlings under salt stress.BMC Plant Biol. 2023 Aug 14;23(1):392. doi: 10.1186/s12870-023-04393-7. BMC Plant Biol. 2023. PMID: 37580686 Free PMC article.
-
Identification of EXPA4 as a key gene in cotton salt stress adaptation through transcriptomic and coexpression network analysis of root tip protoplasts.BMC Plant Biol. 2025 Jan 16;25(1):65. doi: 10.1186/s12870-024-05958-w. BMC Plant Biol. 2025. PMID: 39815183 Free PMC article.
References
LinkOut - more resources
Full Text Sources

