Deciphering the core seed endo-bacteriome of the highland barley in Tibet plateau
- PMID: 36388601
- PMCID: PMC9650301
- DOI: 10.3389/fpls.2022.1041504
Deciphering the core seed endo-bacteriome of the highland barley in Tibet plateau
Abstract
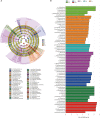
Highland barley (Hordeum vulgare var. nudum (L.) Hook.f., qingke) has unique physical and chemical properties and good potential for industrial applications. As the only crop that can be grown at high altitudes of 4200-4500 m, qingke is well adapted to extreme habitats at high altitudes. In this study, we analysed the seed bacterial community of 58 genotypes of qingke grown in different regions of Tibet, including qingke landraces, modern cultivars, and winter barley varieties, and characterised endophytic bacterial communities in seeds from different sources and the core endo-bacteriome of qingke. This study aim to provide a reference for the application of seed endophytes as biological inoculants for sustainable agricultural production and for considering microbe-plant interactions in breeding strategies. A total of 174 qingke seed samples from five main agricultural regions in Tibet were collected and subjected to investigation of endophytic endo-bacteriome using high-throughput sequencing and bioinformatics approaches. The phyla of endophytic bacteria in qingke seeds from different sources were similar; however, the relative proportions of each phylum were different. Different environmental conditions, growth strategies, and modern breeding processes have significantly changed the community structure of endophytic bacteria in seeds, among which the growth strategy has a greater impact on the diversity of endophytic bacteria in seeds. Seeds from different sources have conserved beneficial core endo-bacteriome. The core endo-bacteriome of qingke seeds dominated by Enterobacteriaceae may maintain qingke growth by promoting plant growth and assisting plants in resisting pests and diseases. This study reveals the core endo-bacteriome of qingke seeds and provides a basis for exploiting the endophytic endo-bacteriome of qingke seeds.
Keywords: core endo-bacteriome; endophytes; highland barley; qingke; seed.
Copyright © 2022 Hao, Wang, Guo and De.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures
Similar articles
-
Characterization of Genetic Diversity and Genome-Wide Association Mapping of Three Agronomic Traits in Qingke Barley (Hordeum Vulgare L.) in the Qinghai-Tibet Plateau.Front Genet. 2020 Jul 3;11:638. doi: 10.3389/fgene.2020.00638. eCollection 2020. Front Genet. 2020. PMID: 32719715 Free PMC article.
-
High level of conservation and diversity among the endophytic seed bacteriome in eight alpine grassland species growing at the Qinghai Tibetan Plateau.FEMS Microbiol Ecol. 2021 May 25;97(6):fiab060. doi: 10.1093/femsec/fiab060. FEMS Microbiol Ecol. 2021. PMID: 33885767
-
The treasure inside barley seeds: microbial diversity and plant beneficial bacteria.Environ Microbiome. 2021 Oct 28;16(1):20. doi: 10.1186/s40793-021-00389-8. Environ Microbiome. 2021. PMID: 34711269 Free PMC article.
-
Highland barley starch (Qingke): Structures, properties, modifications, and applications.Int J Biol Macromol. 2021 Aug 31;185:725-738. doi: 10.1016/j.ijbiomac.2021.06.204. Epub 2021 Jul 2. Int J Biol Macromol. 2021. PMID: 34224757 Review.
-
Plant probiotics - Endophytes pivotal to plant health.Microbiol Res. 2022 Oct;263:127148. doi: 10.1016/j.micres.2022.127148. Epub 2022 Jul 27. Microbiol Res. 2022. PMID: 35940110 Review.
References
-
- Arafat Y., Ud Din I., Tayyab M., Jiang Y., Chen T., Cai Z., et al. . (2020). Soil sickness in aged tea plantation is associated with a shift in microbial communities as a result of plant polyphenol accumulation in the tea gardens. Front. Plant Sci. 11. doi: 10.3389/fpls.2020.00601 - DOI - PMC - PubMed
-
- Bergna A., Cernava T., Rändler M., Grosch R., Zachow C., Berg G. (2018). Tomato seeds preferably transmit plant beneficial endophytes. Phytobiomes. J. 2, 183–193. doi: 10.1094/PBIOMES-06-18-0029-R - DOI
LinkOut - more resources
Full Text Sources
Research Materials

