Uganda Genome Resource: A rich research database for genomic studies of communicable and non-communicable diseases in Africa
- PMID: 36388767
- PMCID: PMC9646479
- DOI: 10.1016/j.xgen.2022.100209
Uganda Genome Resource: A rich research database for genomic studies of communicable and non-communicable diseases in Africa
Abstract
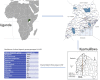
The Uganda Genome Resource (UGR) is a well-characterized genomic database with a range of phenotypic communicable and non-communicable diseases and risk factors generated from the Uganda General Population Cohort (GPC), a population-based open cohort established in 1989. The UGR comprises genotype data on ∼5,000 and whole-genome sequence data on ∼2,000 Ugandan GPC individuals from 10 ethno-linguistic groups. Leveraging other platforms at MRC/UVRI and LSHTM Uganda Research Unit, there is opportunity for additional sample collection to expand the UGR to advance scientific discoveries. Here, we describe UGR and highlight how it is providing opportunities for discovery of novel disease susceptibility genetic loci, refining association signals at new and existing loci, developing and testing polygenic scores to determine disease risk, assessing causal relations in diseases, and developing capacity for genomics research in Africa. The UGR has the potential to develop to a comparable level of European and Asian large-scale genomic initiatives.
Keywords: GPC; NCDs; Uganda; general population cohort; genomics; non-communicable diseases.
© 2022 The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures
References
-
- Asiki G., Murphy G., Nakiyingi-Miiro J., Seeley J., Nsubuga R.N., Karabarinde A., Waswa L., Biraro S., Kasamba I., Pomilla C., et al. GPC team The general population cohort in rural south-western Uganda: a platform for communicable and non-communicable disease studies. Int. J. Epidemiol. 2013;42:129–141. doi: 10.1093/ije/dys234. - DOI - PMC - PubMed
-
- Gurdasani D., Carstensen T., Fatumo S., Chen G., Franklin C.S., Prado-Martinez J., Bouman H., Abascal F., Haber M., Tachmazidou I., et al. Uganda genome resource enables insights into population history and genomic discovery in Africa. Cell. 2019;179:984–1002.e36. doi: 10.1016/j.cell.2019.10.004. - DOI - PMC - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources