Evaluation of reference genes for transcript normalization in Fragaria chiloensis fruit and vegetative tissues
- PMID: 36389093
- PMCID: PMC9530087
- DOI: 10.1007/s12298-022-01227-y
Evaluation of reference genes for transcript normalization in Fragaria chiloensis fruit and vegetative tissues
Abstract
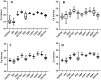
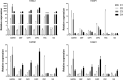
Quantitative real-time PCR (RT-qPCR) is used extensively in gene expression studies. For adequate comparisons, the identification and use of reliable reference genes are crucial. Nevertheless, the availability of such genes in strawberry species is limited and has yet to be described for the Chilean strawberry, Fragaria chiloensis. In this study, the expression dynamics of a set of 10 candidate reference genes were analyzed in various F. chiloensis vegetative tissues (root, runners, stem, leaf, and flower), and fruits at different ripening stages or subjected to different hormonal treatments (ABA, auxin). The expression stability of candidate genes was examined by a series of algorithms, such as geNorm, NormFinder, BestKeeper, and ΔCt, for comparisons and rankings. Finally, by using RefFinder, a comprehensive and comparative ranking of the four methods was achieved. The results highlight that the expression stability of candidate reference genes fluctuates depending on tissue type, fruit stage, and hormonal treatment. As reference genes, the use of FcCHP2 and FcACTIN1 is recommended for F. chiloensis vegetative tissues; FcDBP and FcCHP1 for fruit ripening stages; FcGAPDH and FcDBP for fruit subjected to ABA and NDGA treatments; FcCHP1 and FcCHP2 for fruit under AUXIN and TIBA treatments; and FcDBP and FcCHP2 when all fruit stages and hormonal treatments are compared. If just one reference gene is employed as a normalizer, FcDBP should be chosen as it is the most stable internal control in most conditions. Therefore, the present study delivers a set of reliable reference genes for RT-qPCR expression analysis in F. chiloensis tissues and fruits subjected to several hormonal treatments.
Supplementary information: The online version contains supplementary material available at 10.1007/s12298-022-01227-y.
Keywords: Fragaria chiloensis; Fruit ripening; Normalization; RT-qPCR; Reference genes.
© Prof. H.S. Srivastava Foundation for Science and Society 2022, Springer Nature or its licensor holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
Conflict of interest statement
Conflict of interestThe authors have no relevant financial or non-financial interests to disclose.
Figures
References
-
- Amil-Ruiz F, Garrido-Gala J, Blanco-Portales R, Folta KM, Muñoz-Blanco J, Caballero JL. Identification and validation of reference genes for transcript normalization in strawberry (Fragaria × ananassa) defense responses. PLoS ONE. 2013;8:e70603. doi: 10.1371/journal.pone.0070603. - DOI - PMC - PubMed
-
- Andersen CL, Jensen JL, Ørntoft TF. Normalization of real-time quantitative reverse transcription-PCR data: a model-based variance estimation approach to identify genes suited for normalization, applied to bladder and colon cancer data sets. Cancer Res. 2004;64:5245–5250. doi: 10.1158/0008-5472.CAN-04-0496. - DOI - PubMed
LinkOut - more resources
Full Text Sources
Research Materials
