Know when and how to die: gaining insights into the molecular regulation of leaf senescence
- PMID: 36389097
- PMCID: PMC9530073
- DOI: 10.1007/s12298-022-01224-1
Know when and how to die: gaining insights into the molecular regulation of leaf senescence
Abstract
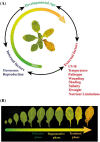
Senescence is the ultimate phase in the life cycle of leaves which is crucial for recycling of nutrients to maintain plant fitness and reproductive success. The earliest visible manifestation of leaf senescence is their yellowing, which usually commences with the breakdown of chlorophyll. The degradation process involves a gradual and highly coordinated disassembly of macromolecules resulting in the accumulation of nutrients, which are subsequently mobilized from the senescing leaves to the developing organs. Leaf senescence progresses under overly tight genetic and molecular control involving a well-orchestrated and intricate network of regulators that coordinate spatio-temporally with the influence of both internal and external cues. Owing to the advancements in omics technologies, the availability of mutant resources, scalability of molecular analyses methodologies and the advanced capacity to integrate multidimensional data, our understanding of the genetic and molecular basis of leaf ageing has greatly expanded. The review provides a compilation of the multitier regulation of senescence process and the interrelation between the environment and the terminal phase of leaf development. The knowledge gained would benefit in devising the strategies for manipulation of leaf senescence process to improve crop quality and productivity.
Keywords: Crop productivity; Omics technologies; Regulation; Senescence.
© Prof. H.S. Srivastava Foundation for Science and Society 2022, Springer Nature or its licensor holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
Conflict of interest statement
Conflict of interestNo competing interest.
Figures

References
Publication types
LinkOut - more resources
Full Text Sources
