New multiple sclerosis lesion segmentation and detection using pre-activation U-Net
- PMID: 36389254
- PMCID: PMC9646406
- DOI: 10.3389/fnins.2022.975862
New multiple sclerosis lesion segmentation and detection using pre-activation U-Net
Abstract
Automated segmentation of new multiple sclerosis (MS) lesions in 3D MRI data is an essential prerequisite for monitoring and quantifying MS progression. Manual delineation of such lesions is time-consuming and expensive, especially because raters need to deal with 3D images and several modalities. In this paper, we propose Pre-U-Net, a 3D encoder-decoder architecture with pre-activation residual blocks, for the segmentation and detection of new MS lesions. Due to the limited training set and the class imbalance problem, we apply intensive data augmentation and use deep supervision to train our models effectively. Following the same U-shaped architecture but different blocks, Pre-U-Net outperforms U-Net and Res-U-Net on the MSSEG-2 dataset, achieving a Dice score of 40.3% on new lesion segmentation and an F1 score of 48.1% on new lesion detection. The codes and trained models are publicly available at https://github.com/pashtari/xunet.
Keywords: U-Net; multiple sclerosis; new lesions; pre-activation; segmentation.
Copyright © 2022 Ashtari, Barile, Van Huffel and Sappey-Marinier.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
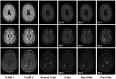
Figures



References
-
- Ashtari P., Barile B., Van Huffel S., Sappey-Marinier D. (2021a). “Longitudinal multiple sclerosis lesion segmentation using pre-activation U-Net,” in MSSEG-2 Challenge Proceedings: Multiple Sclerosis New Lesions Segmentation Challenge Using a Data Management and Processing Infrastructure, (Strasbourg), 45–51. Available online at: https://hal.inria.fr/hal-03358968v1/document#page=54
-
- Ashtari P., Maes F., Van Huffel S. (2021b). Low-rank convolutional networks for brain tumor segmentation, in International MICCAI Brainlesion Workshop: BrainLes 2020. Lecture Notes in Computer Science, Vol. 12658, eds Crimi A., Bakas S. (Cham: Springer; ), 470–480. 10.1007/978-3-030-72084-1_42 - DOI
-
- Çiçek Ö., Abdulkadir A., Lienkamp S. S., Brox T., Ronneberger O. (2016). 3D U-Net: learning dense volumetric segmentation from sparse annotation, in Medical Image Computing and Computer-Assisted Intervention-MICCAI 2016, eds Ourselin S., Joskowicz L., Sabuncu M. R., Unal G., Wells W. (Cham: Springer; ), 424–432. 10.1007/978-3-319-46723-8_49 - DOI
LinkOut - more resources
Full Text Sources

