CD8+ T-cell immune escape by SARS-CoV-2 variants of concern
- PMID: 36389664
- PMCID: PMC9647062
- DOI: 10.3389/fimmu.2022.962079
CD8+ T-cell immune escape by SARS-CoV-2 variants of concern
Abstract
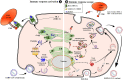
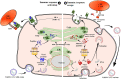
Despite the efficacy of antiviral drug repositioning, convalescent plasma (CP), and the currently available vaccines against severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), the worldwide coronavirus disease 2019 (COVID-19) pandemic is still challenging because of the ongoing emergence of certain new SARS-CoV-2 strains known as variants of concern (VOCs). Mutations occurring within the viral genome, characterized by these new emerging VOCs, confer on them the ability to efficiently resist and escape natural and vaccine-induced humoral and cellular immune responses. Consequently, these VOCs have enhanced infectivity, increasing their stable spread in a given population with an important fatality rate. While the humoral immune escape process is well documented, the evasion mechanisms of VOCs from cellular immunity are not well elaborated. In this review, we discussed how SARS-CoV-2 VOCs adapt inside host cells and escape anti-COVID-19 cellular immunity, focusing on the effect of specific SARS-CoV-2 mutations in hampering the activation of CD8+ T-cell immunity.
Keywords: CD8 + T-cell epitope; HLA; SARS-CoV-2; cellular immunity; cytotoxic T lymphocytes (CTL); immune escape; protein mutation; variant of concern (VOC).
Copyright © 2022 Kombe Kombe, Biteghe, Ndoutoume and Jin.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures


Similar articles
-
Structural insights into immune escape at killer T cell epitope by SARS-CoV-2 Spike Y453F variants.J Biol Chem. 2024 Aug;300(8):107563. doi: 10.1016/j.jbc.2024.107563. Epub 2024 Jul 11. J Biol Chem. 2024. PMID: 39002680 Free PMC article.
-
Cross-protection induced by highly conserved human B, CD4+, and CD8+ T-cell epitopes-based vaccine against severe infection, disease, and death caused by multiple SARS-CoV-2 variants of concern.Front Immunol. 2024 Jan 22;15:1328905. doi: 10.3389/fimmu.2024.1328905. eCollection 2024. Front Immunol. 2024. PMID: 38318166 Free PMC article.
-
CD8+ T-Cell Epitope Variations Suggest a Potential Antigen HLA-A2 Binding Deficiency for Spike Protein of SARS-CoV-2.Front Immunol. 2022 Jan 18;12:764949. doi: 10.3389/fimmu.2021.764949. eCollection 2021. Front Immunol. 2022. PMID: 35116022 Free PMC article.
-
Humoral and cellular immunity against diverse SARS-CoV-2 variants.J Genet Genomics. 2023 Dec;50(12):934-947. doi: 10.1016/j.jgg.2023.10.003. Epub 2023 Oct 19. J Genet Genomics. 2023. PMID: 37865193 Review.
-
Omicron: What Makes the Latest SARS-CoV-2 Variant of Concern So Concerning?J Virol. 2022 Mar 23;96(6):e0207721. doi: 10.1128/jvi.02077-21. Epub 2022 Mar 23. J Virol. 2022. PMID: 35225672 Free PMC article. Review.
Cited by
-
Broad protective RBD heterotrimer vaccines neutralize SARS-CoV-2 including Omicron sub-variants XBB/BQ.1.1/BF.7.PLoS Pathog. 2023 Sep 18;19(9):e1011659. doi: 10.1371/journal.ppat.1011659. eCollection 2023 Sep. PLoS Pathog. 2023. PMID: 37721934 Free PMC article.
-
The molecular mechanisms of CD8+ T cell responses to SARS-CoV-2 infection mediated by TCR-pMHC interactions.Front Immunol. 2024 Oct 10;15:1468456. doi: 10.3389/fimmu.2024.1468456. eCollection 2024. Front Immunol. 2024. PMID: 39450171 Free PMC article. Review.
-
Breadth and Durability of SARS-CoV-2-Specific T Cell Responses following Long-Term Recovery from COVID-19.Microbiol Spectr. 2023 Aug 17;11(4):e0214323. doi: 10.1128/spectrum.02143-23. Epub 2023 Jul 10. Microbiol Spectr. 2023. PMID: 37428088 Free PMC article.
-
The trend of phylogenetic and epitope variations of SARS-CoV-2 Omicron sub-lineages in Iran.Front Microbiol. 2025 Jan 29;15:1531712. doi: 10.3389/fmicb.2024.1531712. eCollection 2024. Front Microbiol. 2025. PMID: 39943963 Free PMC article.
-
Interplay of TLR4 and SARS-CoV-2: Unveiling the Complex Mechanisms of Inflammation and Severity in COVID-19 Infections.J Inflamm Res. 2024 Jul 26;17:5077-5091. doi: 10.2147/JIR.S474707. eCollection 2024. J Inflamm Res. 2024. PMID: 39081874 Free PMC article. Review.
References
-
- Aleem A, Akbar Samad AB, Slenker AK. Emerging variants of SARS-CoV-2 and novel therapeutics against coronavirus (COVID-19). In: StatPearls. FL: Treasure Island; (2022). - PubMed
Publication types
MeSH terms
Supplementary concepts
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

