Early pathogenesis profiles across SARS-CoV-2 variants in K18-hACE2 mice revealed differential triggers of lung damages
- PMID: 36389747
- PMCID: PMC9648130
- DOI: 10.3389/fimmu.2022.950666
Early pathogenesis profiles across SARS-CoV-2 variants in K18-hACE2 mice revealed differential triggers of lung damages
Abstract
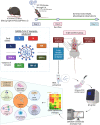
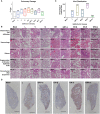
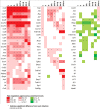
The on-going COVID-19 pandemic has given rise to SARS-CoV-2 clades and variants with differing levels of symptoms and severity. To this end, we aim to systematically elucidate the changes in the pathogenesis as SARS-CoV-2 evolved from ancestral to the recent Omicron VOC, on their mechanisms (e.g. cytokine storm) resulting in tissue damage, using the established K18-hACE2 murine model. We reported that among the SARS-CoV-2 viruses tested, infection profiles were initially similar between viruses from early clades but started to differ greatly starting from VOC Delta, where the trend continues in Omicron. VOCs Delta and Omicron both accumulated a significant number of mutations, and when compared to VOCs Alpha, Beta, and earlier predecessors, showed reduced neurotropism and less apparent gene expression in cytokine storm associated pathways. They were shown to leverage on other pathways to cause tissue damage (or lack of in the case of Omicron). Our study highlighted the importance of elucidating the response profiles of individual SARS-CoV-2 iterations, as their propensity of severe infection via pathways like cytokine storm changes as more variant evolves. This will then affect the overall threat assessment of each variant as well as the use of immunomodulatory treatments as management of severe infections of each variant.
Keywords: K18-hACE2 mice model; SARS-CoV-2; cytokine storm; immune response; variants of concern.
Copyright © 2022 Aw, Mok, Wong, Chen, Mak, Lin, Lye, Tan and Chu.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures


Similar articles
-
The Delta SARS-CoV-2 Variant of Concern Induces Distinct Pathogenic Patterns of Respiratory Disease in K18-hACE2 Transgenic Mice Compared to the Ancestral Strain from Wuhan.mBio. 2022 Jun 28;13(3):e0068322. doi: 10.1128/mbio.00683-22. Epub 2022 Apr 14. mBio. 2022. PMID: 35420469 Free PMC article.
-
Infectious Clones Produce SARS-CoV-2 That Causes Severe Pulmonary Disease in Infected K18-Human ACE2 Mice.mBio. 2021 Apr 20;12(2):e00819-21. doi: 10.1128/mBio.00819-21. mBio. 2021. PMID: 33879586 Free PMC article.
-
Comparison of the Pathogenicity of SARS-CoV-2 Delta and Omicron Variants by Analyzing the Expression Patterns of Immune Response Genes in K18-hACE2 Transgenic Mice.Front Biosci (Landmark Ed). 2022 Nov 30;27(11):316. doi: 10.31083/j.fbl2711316. Front Biosci (Landmark Ed). 2022. PMID: 36472114
-
SARS-CoV-2 Variants of Concern.Yonsei Med J. 2021 Nov;62(11):961-968. doi: 10.3349/ymj.2021.62.11.961. Yonsei Med J. 2021. PMID: 34672129 Free PMC article. Review.
-
Molecular Pathogenesis of Fibrosis, Thrombosis and Surfactant Dysfunction in the Lungs of Severe COVID-19 Patients.Biomolecules. 2022 Dec 10;12(12):1845. doi: 10.3390/biom12121845. Biomolecules. 2022. PMID: 36551272 Free PMC article. Review.
Cited by
-
Neurobiological Alterations Induced by SARS-CoV-2: Insights from Variant-Specific Host Gene Expression Patterns in hACE2-Expressing Mice.Viruses. 2025 Feb 27;17(3):329. doi: 10.3390/v17030329. Viruses. 2025. PMID: 40143258 Free PMC article.
-
Upregulation of inflammatory genes and pathways links obesity to severe COVID-19.Biochim Biophys Acta Mol Basis Dis. 2024 Oct;1870(7):167322. doi: 10.1016/j.bbadis.2024.167322. Epub 2024 Jun 26. Biochim Biophys Acta Mol Basis Dis. 2024. PMID: 38942338 Free PMC article.
-
Spatiotemporal Characterization of Changes in the Respiratory Tract and the Nervous System, Including the Eyes in SARS-CoV-2-Infected K18-hACE2 Mice.Viruses. 2025 Jul 9;17(7):963. doi: 10.3390/v17070963. Viruses. 2025. PMID: 40733579 Free PMC article.
-
Severe Acute Respiratory Syndrome Coronavirus 2 Variant Infection Dynamics and Pathogenesis in Transgenic K18-hACE2 and Inbred Immunocompetent C57BL/6J Mice.Viruses. 2025 Mar 30;17(4):500. doi: 10.3390/v17040500. Viruses. 2025. PMID: 40284943 Free PMC article.
References
Publication types
MeSH terms
Substances
Supplementary concepts
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

